Fowlpox virus encodes a novel DNA repair enzyme, CPD-photolyase, that restores infectivity of UV light-damaged virus
- PMID: 11160666
- PMCID: PMC114077
- DOI: 10.1128/JVI.75.4.1681-1688.2001
Fowlpox virus encodes a novel DNA repair enzyme, CPD-photolyase, that restores infectivity of UV light-damaged virus
Abstract
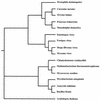
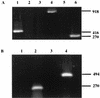
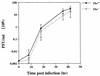
Fowlpox virus (FPV), a pathogen of poultry, can persist in desiccated scabs shed from infected hosts. Although the mechanisms which ensure virus survival are unknown, it is likely that some type of remedial action against environmentally induced damage is required. In this regard, we have identified an open reading frame (ORF) coding for a putative class II cyclobutane pyrimidine dimer (CPD)-photolyase in the genome of FPV. This enzyme repairs the UV light-induced formation of CPDs in DNA by using blue light as an energy source and thus could enhance the viability of FPV during its exposure to sunlight. Based on transcriptional analyses, the photolyase gene was found to be expressed late during the FPV replicative cycle. That the resultant protein retained DNA repair activity was demonstrated by the ability of the corresponding FPV ORF to complement functionally a photolyase-deficient Escherichia coli strain. Interestingly, insertional inactivation of the FPV photolyase gene did not impair the replication of such a genetically altered virus in cultured cells. However, greater sensitivity of this mutant than of the parental virus to UV light irradiation was evident when both were subsequently photoreactivated in the absence of host participation. Therefore, FPV appears to incorporate its photolyase into mature virions where the enzyme can promote their survival in the environment. Although expression of a homologous protein has been predicted for some chordopoxviruses, this report is the first to demonstrate that a poxvirus can utilize light to repair damage to its genome.
Figures



Similar articles
-
The DNA repair enzyme, CPD-photolyase restores the infectivity of UV-damaged fowlpox virus isolated from infected scabs of chickens.Vet Microbiol. 2005 Jul 1;108(3-4):215-23. doi: 10.1016/j.vetmic.2005.04.015. Vet Microbiol. 2005. PMID: 15936904
-
Genetic and phylogenetic characterization of the type II cyclobutane pyrimidine dimer photolyases encoded by Leporipoxviruses.Virology. 2003 Oct 10;315(1):10-9. doi: 10.1016/s0042-6822(03)00512-9. Virology. 2003. PMID: 14592755
-
The genome of fowlpox virus.J Virol. 2000 Apr;74(8):3815-31. doi: 10.1128/jvi.74.8.3815-3831.2000. J Virol. 2000. PMID: 10729156 Free PMC article.
-
Photolyase Production and Current Applications: A Review.Molecules. 2022 Sep 15;27(18):5998. doi: 10.3390/molecules27185998. Molecules. 2022. PMID: 36144740 Free PMC article. Review.
-
Repair of (6-4) Lesions in DNA by (6-4) Photolyase: 20 Years of Quest for the Photoreaction Mechanism.Photochem Photobiol. 2017 Jan;93(1):51-66. doi: 10.1111/php.12696. Epub 2017 Jan 31. Photochem Photobiol. 2017. PMID: 27992654 Review.
Cited by
-
Evaluation of the effectiveness of the SurePure Turbulator ultraviolet-C irradiation equipment on inactivation of different enveloped and non-enveloped viruses inoculated in commercially collected liquid animal plasma.PLoS One. 2019 Feb 21;14(2):e0212332. doi: 10.1371/journal.pone.0212332. eCollection 2019. PLoS One. 2019. PMID: 30789926 Free PMC article.
-
Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair.J Nucleic Acids. 2010 Dec 16;2010:592980. doi: 10.4061/2010/592980. J Nucleic Acids. 2010. PMID: 21209706 Free PMC article.
-
Inactivation of Foodborne Viruses by UV Light: A Review.Foods. 2021 Dec 18;10(12):3141. doi: 10.3390/foods10123141. Foods. 2021. PMID: 34945692 Free PMC article. Review.
-
Molecular Mechanisms of Poxvirus Evolution.mBio. 2023 Feb 28;14(1):e0152622. doi: 10.1128/mbio.01526-22. Epub 2022 Dec 14. mBio. 2023. PMID: 36515529 Free PMC article. Review.
-
An efficient method for generating poxvirus recombinants in the absence of selection.Viruses. 2011 Mar;3(3):217-32. doi: 10.3390/v3030217. Viruses. 2011. PMID: 21494427 Free PMC article.
References
-
- Adachi J, Hasegawa M. Molphy version 2.3 programs for molecular phylogenetics based on maximum likelihood. Computer Science Monographs no. 28. Tokyo, Japan: Institute of Statistical Methods; 1996.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

