Reactivation of the human cytomegalovirus major immediate-early regulatory region and viral replication in embryonal NTera2 cells: role of trichostatin A, retinoic acid, and deletion of the 21-base-pair repeats and modulator
- PMID: 11160656
- PMCID: PMC114067
- DOI: 10.1128/JVI.75.4.1581-1593.2001
Reactivation of the human cytomegalovirus major immediate-early regulatory region and viral replication in embryonal NTera2 cells: role of trichostatin A, retinoic acid, and deletion of the 21-base-pair repeats and modulator
Abstract
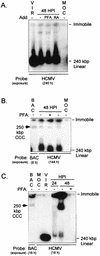
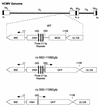
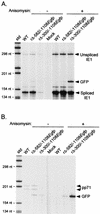
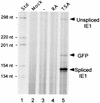
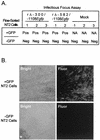
Inactivity of the human cytomegalovirus (HCMV) major immediate-early regulatory region (MIERR), which is composed of promoter, enhancer, unique region, and modulator, is linked to lack of HCMV replication in latently infected cells and in other nonpermissive cell types, including human embryonal NTera2 carcinoma (NT2) cells. I refined the embryonal NT2 cell model to enable characterization of the unknown mechanistic basis for silencing of HCMV MIERR-dependent transcription and viral replication in nonpermissive cells. These infected NT2 cells contain nonreplicating viral genomes with electrophoretic mobility equivalent to a supercoiled, bacterial artificial chromosome of comparable molecular weight. MIERR-dependent transcription is minimal to negligible. Increasing the availability of positive-acting transcription factors by retinoic acid (RA) treatment after infection is largely insufficient in reactivating the MIERR. In contrast, trichostatin A (TSA), a histone deacetylase inhibitor, reactivates MIERR-dependent transcription. Contrary to prior findings produced from transfected MIERR segments, deletion of the 21-bp repeats and modulator from the MIERR in the viral genome does not relieve MIERR silencing. To demonstrate that MIERR silencing likely results from enhancer inactivity, I examined an HCMV with a heterologous MIERR promoter that is enhancer dependent but exempt from IE2 p86-mediated negative autoregulation. This heterologous promoter, like its neighboring native MIERR promoter, exhibits immediate-early transcriptional kinetics in fibroblasts. In embryonal NT2 cells, the heterologous MIERR promoter is transcriptionally inactive. This silence is relieved by TSA but not by RA. Remarkably, TSA-induced reactivation of MIERR-dependent transcription from quiescent viral genomes is followed by release of infectious virus. I conclude that a mechanism of active repression imposes a block to MIERR-dependent transcription and viral replication in embryonal NT2 cells. Because TSA overcomes the block, viral gene silencing may involve histone deacetylase-based modification of viral chromatin, which might account for the covalently closed circular conformation of quiescent HCMV genomes.
Figures
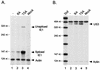

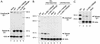

Similar articles
-
Human cytomegalovirus latency is associated with the state of differentiation of the host cells: an in vitro model in teratocarcinoma cells.Acta Microbiol Immunol Hung. 2005;52(3-4):397-406. doi: 10.1556/AMicr.52.2005.3-4.11. Acta Microbiol Immunol Hung. 2005. PMID: 16400879 Review.
-
Breaking human cytomegalovirus major immediate-early gene silence by vasoactive intestinal peptide stimulation of the protein kinase A-CREB-TORC2 signaling cascade in human pluripotent embryonal NTera2 cells.J Virol. 2009 Jul;83(13):6391-403. doi: 10.1128/JVI.00061-09. Epub 2009 Apr 15. J Virol. 2009. PMID: 19369332 Free PMC article.
-
The transcription factor YY1 binds to negative regulatory elements in the human cytomegalovirus major immediate early enhancer/promoter and mediates repression in non-permissive cells.Nucleic Acids Res. 1994 Jul 11;22(13):2453-9. doi: 10.1093/nar/22.13.2453. Nucleic Acids Res. 1994. PMID: 8041605 Free PMC article.
-
The human cytomegalovirus major immediate-early distal enhancer region is required for efficient viral replication and immediate-early gene expression.J Virol. 2000 Feb;74(4):1602-13. doi: 10.1128/jvi.74.4.1602-1613.2000. J Virol. 2000. PMID: 10644329 Free PMC article.
-
Bright and Early: Inhibiting Human Cytomegalovirus by Targeting Major Immediate-Early Gene Expression or Protein Function.Viruses. 2020 Jan 16;12(1):110. doi: 10.3390/v12010110. Viruses. 2020. PMID: 31963209 Free PMC article. Review.
Cited by
-
Transgene expression is associated with copy number and cytomegalovirus promoter methylation in transgenic pigs.PLoS One. 2009 Aug 18;4(8):e6679. doi: 10.1371/journal.pone.0006679. PLoS One. 2009. PMID: 19688097 Free PMC article.
-
Phorbol ester-induced human cytomegalovirus major immediate-early (MIE) enhancer activation through PKC-delta, CREB, and NF-kappaB desilences MIE gene expression in quiescently infected human pluripotent NTera2 cells.J Virol. 2010 Sep;84(17):8495-508. doi: 10.1128/JVI.00416-10. Epub 2010 May 26. J Virol. 2010. PMID: 20504934 Free PMC article.
-
CD8 T cells control cytomegalovirus latency by epitope-specific sensing of transcriptional reactivation.J Virol. 2006 Nov;80(21):10436-56. doi: 10.1128/JVI.01248-06. Epub 2006 Aug 23. J Virol. 2006. PMID: 16928768 Free PMC article.
-
Mouse cytomegalovirus early M112/113 proteins control the repressive effect of IE3 on the major immediate-early promoter.J Virol. 2005 Jan;79(1):257-63. doi: 10.1128/JVI.79.1.257-263.2005. J Virol. 2005. PMID: 15596821 Free PMC article.
-
Polycomb repressive complex 2 silences human cytomegalovirus transcription in quiescent infection models.J Virol. 2013 Dec;87(24):13193-205. doi: 10.1128/JVI.02420-13. Epub 2013 Sep 25. J Virol. 2013. PMID: 24067968 Free PMC article.
References
-
- Andrews P W. Retinoic acid induces neuronal differentiation of a cloned human embryonal carcinoma cell line in vitro. Dev Biol. 1984;103:285–293. - PubMed
-
- Andrews P W, Damjanov I, Simon D, Banting G S, Carlin C, Dracopoli N C, Fogh J. Pluripotent embryonal carcinoma clones derived from the human teratocarcinoma cell line Tera-2. Differentiation in vivo and in vitro. Lab Investig. 1984;50:147–162. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

