The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing
- PMID: 11158314
- PMCID: PMC99581
- DOI: 10.1128/MCB.21.4.1285-1296.2001
The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing
Abstract
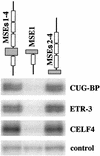
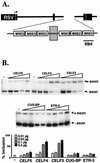
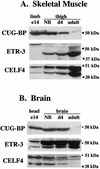
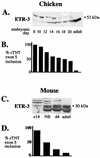
Alternative splicing of cardiac troponin T (cTNT) exon 5 undergoes a developmentally regulated switch such that exon inclusion predominates in embryonic, but not adult, striated muscle. We previously described four muscle-specific splicing enhancers (MSEs) within introns flanking exon 5 in chicken cTNT that are both necessary and sufficient for exon inclusion in embryonic muscle. We also demonstrated that CUG-binding protein (CUG-BP) binds a conserved CUG motif within a human cTNT MSE and positively regulates MSE-dependent exon inclusion. Here we report that CUG-BP is one of a novel family of developmentally regulated RNA binding proteins that includes embryonically lethal abnormal vision-type RNA binding protein 3 (ETR-3). This family, which we call CELF proteins for CUG-BP- and ETR-3-like factors, specifically bound MSE-containing RNAs in vitro and activated MSE-dependent exon inclusion of cTNT minigenes in vivo. The expression of two CELF proteins is highly restricted to brain. CUG-BP, ETR-3, and CELF4 are more broadly expressed, and expression is developmentally regulated in striated muscle and brain. Changes in the level of expression and isoforms of ETR-3 in two different developmental systems correlated with regulated changes in cTNT splicing. A switch from cTNT exon skipping to inclusion tightly correlated with induction of ETR-3 protein expression during differentiation of C2C12 myoblasts. During heart development, the switch in cTNT splicing correlated with a transition in ETR-3 protein isoforms. We propose that ETR-3 is a major regulator of cTNT alternative splicing and that the CELF family plays an important regulatory role in cell-specific alternative splicing during normal development and disease.
Figures


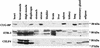


Similar articles
-
Dynamic balance between activation and repression regulates pre-mRNA alternative splicing during heart development.Dev Dyn. 2005 Jul;233(3):783-93. doi: 10.1002/dvdy.20382. Dev Dyn. 2005. PMID: 15830352
-
Muscle-specific splicing enhancers regulate inclusion of the cardiac troponin T alternative exon in embryonic skeletal muscle.Mol Cell Biol. 1996 Aug;16(8):4014-23. doi: 10.1128/MCB.16.8.4014. Mol Cell Biol. 1996. PMID: 8754799 Free PMC article.
-
Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing.Mol Cell. 2002 Mar;9(3):649-58. doi: 10.1016/s1097-2765(02)00479-3. Mol Cell. 2002. PMID: 11931771
-
The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing.Differentiation. 2006 Mar;74(2-3):65-80. doi: 10.1111/j.1432-0436.2006.00060.x. Differentiation. 2006. PMID: 16533306 Review.
-
Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions.Biochimie. 2006 May;88(5):515-25. doi: 10.1016/j.biochi.2005.10.011. Epub 2005 Dec 5. Biochimie. 2006. PMID: 16480813 Review.
Cited by
-
Region-specific alternative splicing in the nervous system: implications for regulation by the RNA-binding protein NAPOR.RNA. 2002 May;8(5):671-85. doi: 10.1017/s1355838202027036. RNA. 2002. PMID: 12022233 Free PMC article.
-
The neurofibromatosis type I pre-mRNA is a novel target of CELF protein-mediated splicing regulation.Nucleic Acids Res. 2010 Jan;38(1):253-64. doi: 10.1093/nar/gkp766. Epub 2009 Oct 23. Nucleic Acids Res. 2010. PMID: 19854948 Free PMC article.
-
Intronic alternative splicing regulators identified by comparative genomics in nematodes.PLoS Comput Biol. 2006 Jul 14;2(7):e86. doi: 10.1371/journal.pcbi.0020086. Epub 2006 Jun 5. PLoS Comput Biol. 2006. PMID: 16839192 Free PMC article.
-
Systematic analysis of cis-elements in unstable mRNAs demonstrates that CUGBP1 is a key regulator of mRNA decay in muscle cells.PLoS One. 2010 Jun 21;5(6):e11201. doi: 10.1371/journal.pone.0011201. PLoS One. 2010. PMID: 20574513 Free PMC article.
-
Flies deficient in Muscleblind protein model features of myotonic dystrophy with altered splice forms of Z-band associated transcripts.Hum Genet. 2006 Nov;120(4):487-99. doi: 10.1007/s00439-006-0228-8. Epub 2006 Aug 23. Hum Genet. 2006. PMID: 16927100
References
-
- Black D L. Activation of c-src neuron-specific splicing by an unusual RNA element in vivo and in vitro. Cell. 1992;69:795–807. - PubMed
-
- Brook J D, McCurrach M E, Harley H G, Buckler A J, Church D, Aburatani H, Hunter K, Stanton V P, Thirion J P, Hudson T, Sohn R, Zemelman B, Snell R G, Rundle S A, Crow S, Davies J, Shelbourne P, Buxton J, Jones C, Juvonen V, Johnson K, Harper P S, Shaw D J, Housman D E. Molecular basis of myotonic dystrophy—expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. - PubMed
-
- Buckanovich R J, Posner J B, Darnell R B. Nova, the paraneoplastic Ri antigen, is homologous to an RNA-binding protein and is specifically expressed in the developing motor system. Neuron. 1993;11:657–672. - PubMed
-
- Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
