hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells
- PMID: 11158309
- PMCID: PMC99576
- DOI: 10.1128/MCB.21.4.1228-1238.2001
hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells
Abstract
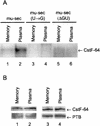
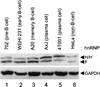
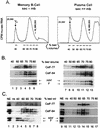
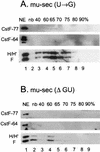
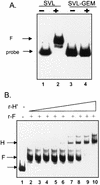
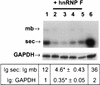
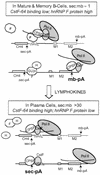
Previous studies on the regulation of polyadenylation of the immunoglobulin (Ig) heavy-chain pre-mRNA argued for trans-acting modifiers of the cleavage-polyadenylation reaction operating differentially during B-cell developmental stages. Using four complementary approaches, we demonstrate that a change in the level of hnRNP F is an important determinant in the regulated use of alternative polyadenylation sites between memory and plasma stage B cells. First, by Western analyses of cellular proteins, the ratio of hnRNP F to H or H' was found to be higher in memory B cells than in plasma cells. In memory B cells the activity of CstF-64 binding to pre-mRNA, but not its amount, was reduced. Second, examination of the complexes formed on input pre-mRNA in nuclear extracts revealed large assemblages containing hnRNP H, H', and F but deficient in CstF-64 in memory B-cell extracts but not in plasma cells. Formation of these large complexes is dependent on the region downstream of the AAUAAA in pre-mRNA, suggesting that CstF-64 and the hnRNPs compete for a similar region. Third, using a recombinant protein we showed that hnRNP F could bind to the region downstream of a poly(A) site, block CstF-64 association with RNA, and inhibit the cleavage reaction. Fourth, overexpression of recombinant hnRNP F in plasma cells resulted in a decrease in the endogenous Ig heavy-chain mRNA secretory form-to-membrane ratio. These results demonstrate that mammalian hnRNP F can act as a negative regulator in the pre-mRNA cleavage reaction and that increased expression of F in memory B cells contributes to the suppression of the Ig heavy-chain secretory poly(A) site.
Figures





Similar articles
-
The binding of a subunit of the general polyadenylation factor cleavage-polyadenylation specificity factor (CPSF) to polyadenylation sites changes during B cell development.Nucleic Acids Symp Ser. 1995;(33):229-33. Nucleic Acids Symp Ser. 1995. PMID: 8643379
-
DSEF-1 is a member of the hnRNP H family of RNA-binding proteins and stimulates pre-mRNA cleavage and polyadenylation in vitro.Nucleic Acids Res. 1998 Dec 1;26(23):5343-50. doi: 10.1093/nar/26.23.5343. Nucleic Acids Res. 1998. PMID: 9826757 Free PMC article.
-
Efficient polyadenylation of Rous sarcoma virus RNA requires the negative regulator of splicing element.Nucleic Acids Res. 2002 Feb 1;30(3):810-7. doi: 10.1093/nar/30.3.810. Nucleic Acids Res. 2002. PMID: 11809895 Free PMC article.
-
A history of poly A sequences: from formation to factors to function.Prog Nucleic Acid Res Mol Biol. 2002;71:285-389. doi: 10.1016/s0079-6603(02)71046-5. Prog Nucleic Acid Res Mol Biol. 2002. PMID: 12102557 Review.
-
Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: Focus on hnRNP E1's multifunctional regulatory roles.RNA. 2010 Aug;16(8):1449-62. doi: 10.1261/rna.2254110. Epub 2010 Jun 28. RNA. 2010. PMID: 20584894 Free PMC article. Review.
Cited by
-
The hnRNP F/H homologue of Trypanosoma brucei is differentially expressed in the two life cycle stages of the parasite and regulates splicing and mRNA stability.Nucleic Acids Res. 2013 Jul;41(13):6577-94. doi: 10.1093/nar/gkt369. Epub 2013 May 10. Nucleic Acids Res. 2013. PMID: 23666624 Free PMC article.
-
The RNA-binding protein hnRNP F is required for the germinal center B cell response.Nat Commun. 2023 Mar 30;14(1):1731. doi: 10.1038/s41467-023-37308-z. Nat Commun. 2023. PMID: 36997512 Free PMC article.
-
Heterogeneity in mammalian RNA 3' end formation.Exp Cell Res. 2010 May 1;316(8):1357-64. doi: 10.1016/j.yexcr.2010.02.040. Epub 2010 Mar 6. Exp Cell Res. 2010. PMID: 20211174 Free PMC article. Review.
-
Biased alternative polyadenylation in human tissues.Genome Biol. 2005;6(12):R100. doi: 10.1186/gb-2005-6-12-r100. Epub 2005 Nov 28. Genome Biol. 2005. PMID: 16356263 Free PMC article.
-
Reconstitution of mammalian cleavage factor II involved in 3' processing of mRNA precursors.RNA. 2018 Dec;24(12):1721-1737. doi: 10.1261/rna.068056.118. Epub 2018 Aug 23. RNA. 2018. PMID: 30139799 Free PMC article.
References
-
- Beyer K, Dandekar T, Keller W. RNA ligands selected by cleavage stimulation factor contain distinct sequence motifs that function as downstream elements in 3′-end processing of pre-mRNA. J Biol Chem. 1997;272:26769–26779. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
