Identification of a plasmid-encoded gene from Haemophilus ducreyi which confers NAD independence
- PMID: 11157928
- PMCID: PMC94989
- DOI: 10.1128/JB.183.4.1168-1174.2001
Identification of a plasmid-encoded gene from Haemophilus ducreyi which confers NAD independence
Abstract
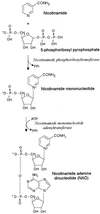
Members of the family Pasteurellaceae are classified in part by whether or not they require an NAD supplement for growth on laboratory media. In this study, we demonstrate that this phenotype can be determined by a single gene, nadV, whose presence allows NAD-independent growth of Haemophilus influenzae and Actinobacillus pleuropneumoniae. This gene was cloned from a 5.2-kb plasmid which was previously shown to be responsible for NAD independence in Haemophilus ducreyi. When transformed into A. pleuropneumoniae, this cloned gene allowed NAD-independent growth on complex media and allowed the utilization of nicotinamide in place of NAD on defined media. Sequence analysis revealed an open reading frame of 1,482 bp that is predicted to encode a protein with a molecular mass of 55,619 Da. Compared with the sequence databases, NadV was found to have significant sequence homology to the human pre-B-cell colony-enhancing factor PBEF and to predicted proteins of unknown function identified in the bacterial species Mycoplasma genitalium, Mycoplasma pneumoniae, Shewanella putrefaciens, Synechocystis sp., Deinococcus radiodurans, Pasteurella multocida, and Actinobacillus actinomycetemcomitans. P. multocida and A. actinomycetemcomitans are among the NAD-independent members of the Pasteurellaceae. Homologues of NadV were not found in the sequenced genome of H. influenzae, an NAD-dependent member of the Pasteurellaceae, or in species known to utilize a different pathway for synthesis of NAD, such as Escherichia coli. Sequence alignment of these nine homologues revealed regions and residues of complete conservation that may be directly involved in the enzymatic activity. Identification of a function for this gene in the Pasteurellaceae should help to elucidate the role of its homologues in other species.
Figures


Similar articles
-
Haemophilus ducreyi strain ATCC 27722 contains a genetic element with homology to the vibrio RS1 element that can replicate as a plasmid and confer NAD independence on haemophilus influenzae.Infect Immun. 2004 Feb;72(2):1143-6. doi: 10.1128/IAI.72.2.1143-1146.2004. Infect Immun. 2004. PMID: 14742562 Free PMC article.
-
Growth characteristics of V factor-independent transformants of Haemophilus influenzae.Int J Syst Bacteriol. 1993 Oct;43(4):799-804. doi: 10.1099/00207713-43-4-799. Int J Syst Bacteriol. 1993. PMID: 8240960
-
An analysis of the complete nucleotide sequence of the Haemophilus ducreyi broad-host-range plasmid pLS88.Plasmid. 1994 Sep;32(2):228-32. doi: 10.1006/plas.1994.1060. Plasmid. 1994. PMID: 7846148
-
Significance of V-factor dependency in the taxonomy of Haemophilus species and related organisms.Int J Syst Bacteriol. 1990 Jan;40(1):1-4. doi: 10.1099/00207713-40-1-1. Int J Syst Bacteriol. 1990. PMID: 2145965 Review. No abstract available.
-
NAD+ utilization in Pasteurellaceae: simplification of a complex pathway.J Bacteriol. 2006 Oct;188(19):6719-27. doi: 10.1128/JB.00432-06. J Bacteriol. 2006. PMID: 16980474 Free PMC article. Review. No abstract available.
Cited by
-
The evolutionary portrait of metazoan NAD salvage.PLoS One. 2013 May 28;8(5):e64674. doi: 10.1371/journal.pone.0064674. Print 2013. PLoS One. 2013. PMID: 23724078 Free PMC article.
-
Nampt is involved in DNA double-strand break repair.Chin J Cancer. 2012 Aug;31(8):392-8. doi: 10.5732/cjc.012.10089. Epub 2012 Jun 14. Chin J Cancer. 2012. PMID: 22704488 Free PMC article.
-
Targeting Danger Signals to Rescue Fibrosis.Am J Respir Cell Mol Biol. 2022 May;66(5):468-470. doi: 10.1165/rcmb.2022-0022ED. Am J Respir Cell Mol Biol. 2022. PMID: 35271415 Free PMC article. No abstract available.
-
Microbial NAD metabolism: lessons from comparative genomics.Microbiol Mol Biol Rev. 2009 Sep;73(3):529-41, Table of Contents. doi: 10.1128/MMBR.00042-08. Microbiol Mol Biol Rev. 2009. PMID: 19721089 Free PMC article. Review.
-
Nicotinamide ribosyl uptake mutants in Haemophilus influenzae.Infect Immun. 2003 Sep;71(9):5398-401. doi: 10.1128/IAI.71.9.5398-5401.2003. Infect Immun. 2003. PMID: 12933892 Free PMC article.
References
-
- Bragg R R, Coetzee L, Verschoor J A. Plasmid-encoded NAD independence in some South African isolates of Haemophilus paragallinarum. Onderstepoort J Vet Res. 1993;60:147–152. - PubMed
-
- Cynamon M H, Sorg T B, Patapow A. Utilization and metabolism of NAD by Haemophilus parainfluenzae. J Gen Microbiol. 1988;134:2789–2799. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

