In situ accessibility of Escherichia coli 23S rRNA to fluorescently labeled oligonucleotide probes
- PMID: 11157269
- PMCID: PMC92673
- DOI: 10.1128/AEM.67.2.961-968.2001
In situ accessibility of Escherichia coli 23S rRNA to fluorescently labeled oligonucleotide probes
Abstract
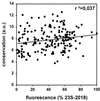
One of the main causes of failure of fluorescence in situ hybridization with rRNA-targeted oligonucleotides, besides low cellular ribosome content and impermeability of cell walls, is the inaccessibility of probe target sites due to higher-order structure of the ribosome. Analogous to a study on the 16S rRNA (B. M. Fuchs, G. Wallner, W. Beisker, I. Schwippl, W. Ludwig, and R. Amann, Appl. Environ. Microbiol. 64:4973-4982, 1998), the accessibility of the 23S rRNA of Escherichia coli DSM 30083(T) was studied in detail with a set of 184 CY3-labeled oligonucleotide probes. The probe-conferred fluorescence was quantified flow cytometrically. The brightest signal resulted from probe 23S-2018, complementary to positions 2018 to 2035. The distribution of probe-conferred cell fluorescence in six arbitrarily set brightness classes (classes I to VI, 100 to 81%, 80 to 61%, 60 to 41%, 40 to 21%, 20 to 6%, and 5 to 0% of the brightness of 23S-2018, respectively) was as follows: class I, 3%; class II, 21%; class III, 35%; class IV, 18%; class V, 16%; and class VI, 7%. A fine-resolution analysis of selected areas confirmed steep changes in accessibility on the 23S RNA to oligonucleotide probes. This is similar to the situation for the 16S rRNA. Indeed, no significant differences were found between the hybridization of oligonucleotide probes to 16S and 23S rRNA. Interestingly, indications were obtained of an effect of the type of fluorescent dye coupled to a probe on in situ accessibility. The results were translated into an accessibility map for the 23S rRNA of E. coli, which may be extrapolated to other bacteria. Thereby, it may contribute to a better exploitation of the high potential of the 23S rRNA for identification of bacteria in the future.
Figures

Similar articles
-
Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes.Appl Environ Microbiol. 1998 Dec;64(12):4973-82. doi: 10.1128/AEM.64.12.4973-4982.1998. Appl Environ Microbiol. 1998. PMID: 9835591 Free PMC article.
-
In situ accessibility of small-subunit rRNA of members of the domains Bacteria, Archaea, and Eucarya to Cy3-labeled oligonucleotide probes.Appl Environ Microbiol. 2003 Mar;69(3):1748-58. doi: 10.1128/AEM.69.3.1748-1758.2003. Appl Environ Microbiol. 2003. PMID: 12620867 Free PMC article.
-
Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms.Cytometry. 1993;14(2):136-43. doi: 10.1002/cyto.990140205. Cytometry. 1993. PMID: 7679962
-
Oligonucleotide probes for RNA-targeted fluorescence in situ hybridization.Adv Clin Chem. 2007;43:79-115. Adv Clin Chem. 2007. PMID: 17249381 Review.
-
Phylogenetic identification and in situ detection of individual microbial cells without cultivation.Microbiol Rev. 1995 Mar;59(1):143-69. doi: 10.1128/mr.59.1.143-169.1995. Microbiol Rev. 1995. PMID: 7535888 Free PMC article. Review.
Cited by
-
Application of Nucleic Acid Mimics in Fluorescence In Situ Hybridization.Methods Mol Biol. 2021;2246:69-86. doi: 10.1007/978-1-0716-1115-9_5. Methods Mol Biol. 2021. PMID: 33576983
-
Development of Fluorescence In Situ Hybridization as a Rapid, Accurate Method for Detecting Coliforms in Water Samples.Biosensors (Basel). 2020 Dec 24;11(1):8. doi: 10.3390/bios11010008. Biosensors (Basel). 2020. PMID: 33374317 Free PMC article.
-
A Transcriptome-Targeting EcoChip for Assessing Functional Mycodiversity.Microarrays (Basel). 2011 Oct 31;1(1):25-41. doi: 10.3390/microarrays1010025. Microarrays (Basel). 2011. PMID: 27605333 Free PMC article.
-
Depletion of unwanted nucleic acid templates by selective cleavage: LNAzymes, catalytically active oligonucleotides containing locked nucleic acids, open a new window for detecting rare microbial community members.Appl Environ Microbiol. 2013 Mar;79(5):1534-44. doi: 10.1128/AEM.03392-12. Epub 2012 Dec 21. Appl Environ Microbiol. 2013. PMID: 23263968 Free PMC article.
-
Advancement of the 10-species subgingival Zurich biofilm model by examining different nutritional conditions and defining the structure of the in vitro biofilms.BMC Microbiol. 2012 Oct 5;12:227. doi: 10.1186/1471-2180-12-227. BMC Microbiol. 2012. PMID: 23040057 Free PMC article.
References
-
- Amann R, Ludwig W. Typing in situ with probes. In: Priest F G, Ramos-Cormenzana A, Tindall B J, editors. Bacterial diversity and systematics. New York, N.Y: Plenum; 1994. pp. 115–135.
-
- Ban N, Nissen P, Hansen J, Moore P B, Steitz T A. The complete atomic structure of the large ribosomal subunit at 2.4 angstrom resolution. Science. 2000;289:905–920. - PubMed
-
- Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

