Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML
- PMID: 11152521
- PMCID: PMC114054
- DOI: 10.1128/JVI.75.3.1487-1506.2001
Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML
Abstract
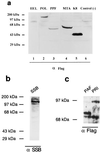
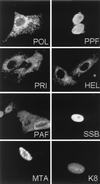
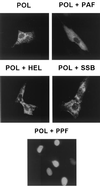
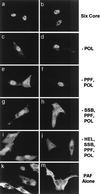
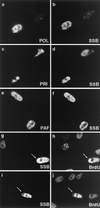
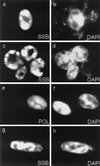
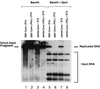
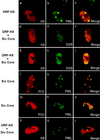
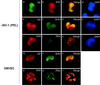
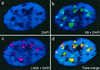
Six predicted Kaposi's sarcoma virus herpesvirus (KSHV) proteins have homology with other well-characterized herpesvirus core DNA replication proteins and are expected to be essential for viral DNA synthesis. Intact Flag-tagged protein products from all six were produced from genomic expression vectors, although the ORF40/41 transcript encoding a primase-helicase component proved to be spliced with a 127-bp intron. The intracellular localization of these six KSHV replication proteins and the mechanism of their nuclear translocation were investigated. SSB (single-stranded DNA binding protein, ORF6) and PPF (polymerase processivity factor, ORF59) were found to be intrinsic nuclear proteins, whereas POL (polymerase, ORF9), which localized in the cytoplasm on its own, was translocated to the nucleus when cotransfected with PPF. PAF (primase-associated factor, ORF40/41), a component of the primase-helicase tripartite subcomplex together with PRI (primase, ORF56) and HEL (helicase, ORF44), required the presence of all five other replication proteins for efficient nuclear translocation. Surprisingly, even in the absence of a lytic cycle replication origin (ori-Lyt) and any known initiator or origin binding protein, the protein products of all six KSHV core replication genes cooperated in a transient cotransfection assay to form large globular shaped pseudo-replication compartments (pseudo-RC), which excluded cellular DNA. These pseudo-RC structures were confirmed to include POL, SSB, PRI, and PAF but did not contain any newly synthesized DNA. Similar to the human cytomegalovirus system, the peripheries of these KSHV pre-RC were also found to be surrounded by punctate PML oncogenic domains (PODs). Furthermore, by transient cotransfection, the six KSHV core replication machinery proteins successfully replicated a plasmid containing EBV ori-Lyt in the presence of the Epstein-Barr virus-encoded DNA binding initiator protein, ZTA. The KSHV-encoded K8 (ORF-K8) protein, which is a distant evolutionary homologue to ZTA, was incorporated into pseudo-RC structures formed by transient cotransfection with the six core KSHV replication genes. However, unlike ZTA, K8 displayed a punctate nuclear pattern both in transfected cells and at early stages of lytic infection and colocalized with the cellular PML proteins in PODs. Finally, K8 was also found to accumulate in functional viral RC, detected by incorporation of pulse-labeled bromodeoxyuridine into newly synthesized DNA in both tetradecanoyl phorbol acetate-induced JSC-1 primary effusion lymphoblasts and in KSHV lytically infected endothelial cells.
Figures



Similar articles
-
Amplification of the Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 lytic origin of DNA replication is dependent upon a cis-acting AT-rich region and an ORF50 response element and the trans-acting factors ORF50 (K-Rta) and K8 (K-bZIP).Virology. 2004 Jan 20;318(2):542-55. doi: 10.1016/j.virol.2003.10.016. Virology. 2004. PMID: 14972523
-
Kaposi's sarcoma-associated herpesvirus lytic origin (ori-Lyt)-dependent DNA replication: identification of the ori-Lyt and association of K8 bZip protein with the origin.J Virol. 2003 May;77(10):5578-88. doi: 10.1128/jvi.77.10.5578-5588.2003. J Virol. 2003. PMID: 12719550 Free PMC article.
-
The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10).J Virol. 1999 Dec;73(12):10458-71. doi: 10.1128/JVI.73.12.10458-10471.1999. J Virol. 1999. PMID: 10559364 Free PMC article.
-
Lytic cycle switches of oncogenic human gammaherpesviruses.Adv Cancer Res. 2007;97:81-109. doi: 10.1016/S0065-230X(06)97004-3. Adv Cancer Res. 2007. PMID: 17419942 Review.
-
Inhibiting KSHV replication by targeting the essential activities of KSHV processivity protein, PF-8.J Med Virol. 2024 Oct;96(10):e29958. doi: 10.1002/jmv.29958. J Med Virol. 2024. PMID: 39370884 Review.
Cited by
-
Gene structure and expression of Kaposi's sarcoma-associated herpesvirus ORF56, ORF57, ORF58, and ORF59.J Virol. 2006 Dec;80(24):11968-81. doi: 10.1128/JVI.01394-06. Epub 2006 Oct 4. J Virol. 2006. PMID: 17020939 Free PMC article.
-
Human cytomegalovirus UL84 insertion mutant defective for viral DNA synthesis and growth.J Virol. 2004 Oct;78(19):10360-9. doi: 10.1128/JVI.78.19.10360-10369.2004. J Virol. 2004. PMID: 15367602 Free PMC article.
-
Disruption of gammaherpesvirus 68 gene 50 demonstrates that Rta is essential for virus replication.J Virol. 2003 May;77(10):5731-9. doi: 10.1128/jvi.77.10.5731-5739.2003. J Virol. 2003. PMID: 12719566 Free PMC article.
-
Potent antiviral activity of north-methanocarbathymidine against Kaposi's sarcoma-associated herpesvirus.Antimicrob Agents Chemother. 2005 Dec;49(12):4965-73. doi: 10.1128/AAC.49.12.4965-4973.2005. Antimicrob Agents Chemother. 2005. PMID: 16304159 Free PMC article.
-
DNA processing by the Kaposi's sarcoma-associated herpesvirus alkaline exonuclease SOX contributes to viral gene expression and infectious virion production.Nucleic Acids Res. 2023 Jan 11;51(1):182-197. doi: 10.1093/nar/gkac1190. Nucleic Acids Res. 2023. PMID: 36537232 Free PMC article.
References
-
- Ballestas M E, Chatis P A, Kaye K M. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

