The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability
- PMID: 11152504
- PMCID: PMC114037
- DOI: 10.1128/JVI.75.3.1312-1324.2001
The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability
Abstract
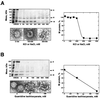
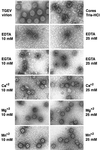
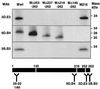
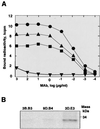
The architecture of transmissible gastroenteritis coronavirus includes three different structural levels, the envelope, an internal core, and the nucleocapsid that is released when the core is disrupted. Starting from purified virions, core structures have been reproducibly isolated as independent entities. The cores were stabilized at basic pH and by the presence of divalent cations, with Mg(2+) ions more effectively contributing to core stability. Core structures showed high resistance to different concentrations of detergents, reducing agents, and urea and low concentrations of monovalent ions (<200 mM). Cores were composed of the nucleoprotein, RNA, and the C domain of the membrane (M) protein. At high salt concentrations (200 to 300 mM), the M protein was no longer associated with the nucleocapsid, which resulted in destruction of the core structure. A specific ionic interaction between the M protein carboxy terminus and the nucleocapsid was demonstrated using three complementary approaches: (i) a binding assay performed between a collection of M protein amino acid substitution or deletion mutants and purified nucleocapsids that led to the identification of a 16-amino-acid (aa) domain (aa 237 to 252) as being responsible for binding the M protein to the nucleocapsid; (ii) the specific inhibition of this binding by monoclonal antibodies (MAbs) binding to a carboxy-terminal M protein domain close to the indicated peptide but not by MAbs specific for the M protein amino terminus; and (iii) a 26-residue peptide, including the predicted sequence (aa 237 to 252), which specifically inhibited the binding. Direct binding of the M protein to the nucleoprotein was predicted, since degradation of the exposed RNA by RNase treatment did not affect the binding. It is proposed that the M protein is embedded within the virus membrane and that the C region, exposed to the interior face of the virion in a population of these molecules, interacts with the nucleocapsid to which it is anchored, forming the core. Only the C region of the M protein is part of the core.
Figures






Similar articles
-
Organization of two transmissible gastroenteritis coronavirus membrane protein topologies within the virion and core.J Virol. 2001 Dec;75(24):12228-40. doi: 10.1128/JVI.75.24.12228-12240.2001. J Virol. 2001. PMID: 11711614 Free PMC article.
-
Membrane protein molecules of transmissible gastroenteritis coronavirus also expose the carboxy-terminal region on the external surface of the virion.J Virol. 1995 Sep;69(9):5269-77. doi: 10.1128/JVI.69.9.5269-5277.1995. J Virol. 1995. PMID: 7636969 Free PMC article.
-
The amino-terminal signal peptide on the porcine transmissible gastroenteritis coronavirus matrix protein is not an absolute requirement for membrane translocation and glycosylation.Virology. 1988 Aug;165(2):367-76. doi: 10.1016/0042-6822(88)90581-8. Virology. 1988. PMID: 2841792 Free PMC article.
-
Coronavirus genomic RNA packaging.Virology. 2019 Nov;537:198-207. doi: 10.1016/j.virol.2019.08.031. Epub 2019 Aug 30. Virology. 2019. PMID: 31505321 Free PMC article. Review.
-
Genetic and molecular biological analysis of protein-protein interactions in coronavirus assembly.Adv Exp Med Biol. 2006;581:163-73. doi: 10.1007/978-0-387-33012-9_29. Adv Exp Med Biol. 2006. PMID: 17037525 Free PMC article. Review. No abstract available.
Cited by
-
Calcium‐dependent antimicrobials: Nature‐inspired materials and designs.Exploration (Beijing). 2024 Mar 12;4(5):20230099. doi: 10.1002/EXP.20230099. eCollection 2024 Oct. Exploration (Beijing). 2024. PMID: 39439493 Free PMC article. Review. Catalan.
-
Humoral Immune Response in Immunized Sheep with Bovine Coronavirus Glycoproteins Delivered via an Adenoviral Vector.Pathogens. 2024 Jun 21;13(7):523. doi: 10.3390/pathogens13070523. Pathogens. 2024. PMID: 39057750 Free PMC article.
-
COVID-19 Variants and Vaccine Development.Viruses. 2024 May 10;16(5):757. doi: 10.3390/v16050757. Viruses. 2024. PMID: 38793638 Free PMC article. Review.
-
SARS-CoV-2 outbreak: role of viral proteins and genomic diversity in virus infection and COVID-19 progression.Virol J. 2024 Mar 27;21(1):75. doi: 10.1186/s12985-024-02342-w. Virol J. 2024. PMID: 38539202 Free PMC article. Review.
-
A Candidate Antigen of the Recombinant Membrane Protein Derived from the Porcine Deltacoronavirus Synthetic Gene to Detect Seropositive Pigs.Viruses. 2023 Apr 25;15(5):1049. doi: 10.3390/v15051049. Viruses. 2023. PMID: 37243136 Free PMC article.
References
-
- Ansorge W. Fast and sensitive detection of protein and DNA bands by treatment with potassium permanganate. J Biochem Biophys Methods. 1985;11:13–20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

