Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS
- PMID: 11148291
- PMCID: PMC102231
- DOI: 10.1105/tpc.12.12.2473
Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS
Abstract
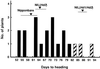
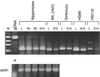
A major quantitative trait locus (QTL) controlling response to photoperiod, Hd1, was identified by means of a map-based cloning strategy. High-resolution mapping using 1505 segregants enabled us to define a genomic region of approximately 12 kb as a candidate for Hd1. Further analysis revealed that the Hd1 QTL corresponds to a gene that is a homolog of CONSTANS in Arabidopsis. Sequencing analysis revealed a 43-bp deletion in the first exon of the photoperiod sensitivity 1 (se1) mutant HS66 and a 433-bp insertion in the intron in mutant HS110. Se1 is allelic to the Hd1 QTL, as determined by analysis of two se1 mutants, HS66 and HS110. Genetic complementation analysis proved the function of the candidate gene. The amount of Hd1 mRNA was not greatly affected by a change in length of the photoperiod. We suggest that Hd1 functions in the promotion of heading under short-day conditions and in inhibition under long-day conditions.
Figures



Comment in
-
Giving rice the time of day. Molecular identification of a major photoperiod sensitivity quantitative trait locus.Plant Cell. 2000 Dec;12(12):2299-2301. doi: 10.1105/tpc.12.12.2299. Plant Cell. 2000. PMID: 11148278 Free PMC article. No abstract available.
Similar articles
-
Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions.Plant Cell Physiol. 2002 Oct;43(10):1096-105. doi: 10.1093/pcp/pcf156. Plant Cell Physiol. 2002. PMID: 12407188
-
Characterization and functional analysis of three wheat genes with homology to the CONSTANS flowering time gene in transgenic rice.Plant J. 2003 Oct;36(1):82-93. doi: 10.1046/j.1365-313x.2003.01859.x. Plant J. 2003. PMID: 12974813
-
The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis.Plant Physiol. 2003 Apr;131(4):1855-67. doi: 10.1104/pp.102.016188. Plant Physiol. 2003. PMID: 12692345 Free PMC article.
-
Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1.Genes Dev. 2004 Apr 15;18(8):926-36. doi: 10.1101/gad.1189604. Epub 2004 Apr 12. Genes Dev. 2004. PMID: 15078816 Free PMC article.
-
[Molecular mechanism controlling daylength regulation of flowering in short-day plants].Tanpakushitsu Kakusan Koso. 2002 Sep;47(12 Suppl):1541-6. Tanpakushitsu Kakusan Koso. 2002. PMID: 12357608 Review. Japanese. No abstract available.
Cited by
-
MITE Digger, an efficient and accurate algorithm for genome wide discovery of miniature inverted repeat transposable elements.BMC Bioinformatics. 2013 Jun 7;14:186. doi: 10.1186/1471-2105-14-186. BMC Bioinformatics. 2013. PMID: 23758809 Free PMC article.
-
Punctual transcriptional regulation by the rice circadian clock under fluctuating field conditions.Plant Cell. 2015 Mar;27(3):633-48. doi: 10.1105/tpc.114.135582. Epub 2015 Mar 10. Plant Cell. 2015. PMID: 25757473 Free PMC article.
-
The rice B-box zinc finger gene family: genomic identification, characterization, expression profiling and diurnal analysis.PLoS One. 2012;7(10):e48242. doi: 10.1371/journal.pone.0048242. Epub 2012 Oct 31. PLoS One. 2012. PMID: 23118960 Free PMC article.
-
Strong photoperiod sensitivity is controlled by cooperation and competition among Hd1, Ghd7 and DTH8 in rice heading.New Phytol. 2021 Feb;229(3):1635-1649. doi: 10.1111/nph.16946. Epub 2020 Oct 22. New Phytol. 2021. PMID: 33089895 Free PMC article.
-
Molecular basis of heading date control in rice.aBIOTECH. 2020 May 11;1(4):219-232. doi: 10.1007/s42994-020-00019-w. eCollection 2020 Oct. aBIOTECH. 2020. PMID: 36304129 Free PMC article. Review.
References
-
- Alonso-Blanco, C., and Koornneef, M. (2000). Naturally occurring variation in Arabidopsis: An underexploited resource for plant genetics. Trends Plant Sci. 5 22–29. - PubMed
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. - PubMed
-
- Baba, T., et al. (2000). Construction and characterization of rice genomic libraries, PAC library of japonica variety Nipponbare, and BAC library of indica variety Kasalath. Bull. Natl. Inst. Agrobiol. Resour. 14 41–49.
-
- Burge, C., and Karlin, S. (1997). Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268 78–94. - PubMed
-
- Chomcznski, P., and Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 162 156–159. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

