Pandemic threat posed by avian influenza A viruses
- PMID: 11148006
- PMCID: PMC88966
- DOI: 10.1128/CMR.14.1.129-149.2001
Pandemic threat posed by avian influenza A viruses
Abstract
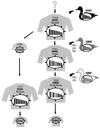
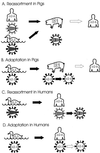
Influenza pandemics, defined as global outbreaks of the disease due to viruses with new antigenic subtypes, have exacted high death tolls from human populations. The last two pandemics were caused by hybrid viruses, or reassortants, that harbored a combination of avian and human viral genes. Avian influenza viruses are therefore key contributors to the emergence of human influenza pandemics. In 1997, an H5N1 influenza virus was directly transmitted from birds in live poultry markets in Hong Kong to humans. Eighteen people were infected in this outbreak, six of whom died. This avian virus exhibited high virulence in both avian and mammalian species, causing systemic infection in both chickens and mice. Subsequently, another avian virus with the H9N2 subtype was directly transmitted from birds to humans in Hong Kong. Interestingly, the genes encoding the internal proteins of the H9N2 virus are genetically highly related to those of the H5N1 virus, suggesting a unique property of these gene products. The identification of avian viruses in humans underscores the potential of these and similar strains to produce devastating influenza outbreaks in major population centers. Although highly pathogenic avian influenza viruses had been identified before the 1997 outbreak in Hong Kong, their devastating effects had been confined to poultry. With the Hong Kong outbreak, it became clear that the virulence potential of these viruses extended to humans.
Figures
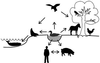


Similar articles
-
Continued circulation in China of highly pathogenic avian influenza viruses encoding the hemagglutinin gene associated with the 1997 H5N1 outbreak in poultry and humans.J Virol. 2000 Jul;74(14):6592-9. doi: 10.1128/jvi.74.14.6592-6599.2000. J Virol. 2000. PMID: 10864673 Free PMC article.
-
The continued pandemic threat posed by avian influenza viruses in Hong Kong.Trends Microbiol. 2002 Jul;10(7):340-4. doi: 10.1016/s0966-842x(02)02388-0. Trends Microbiol. 2002. PMID: 12110213 Review.
-
H9N2 subtype influenza A viruses in poultry in pakistan are closely related to the H9N2 viruses responsible for human infection in Hong Kong.Virology. 2000 Dec 5;278(1):36-41. doi: 10.1006/viro.2000.0585. Virology. 2000. PMID: 11112478
-
Characterization of avian H5N1 influenza viruses from poultry in Hong Kong.Virology. 1998 Dec 20;252(2):331-42. doi: 10.1006/viro.1998.9488. Virology. 1998. PMID: 9878612
-
Avian influenza and human health.Acta Trop. 2002 Jul;83(1):1-6. doi: 10.1016/s0001-706x(02)00050-5. Acta Trop. 2002. PMID: 12062786 Review.
Cited by
-
Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003.J Clin Microbiol. 2005 Nov;43(11):5760-7. doi: 10.1128/JCM.43.11.5760-5767.2005. J Clin Microbiol. 2005. PMID: 16272514 Free PMC article.
-
Influenza infection in wild raccoons.Emerg Infect Dis. 2008 Dec;14(12):1842-8. doi: 10.3201/eid1412.071371. Emerg Infect Dis. 2008. PMID: 19046505 Free PMC article.
-
Alterations in hemagglutinin receptor-binding specificity accompany the emergence of highly pathogenic avian influenza viruses.J Virol. 2015 May;89(10):5395-405. doi: 10.1128/JVI.03304-14. Epub 2015 Mar 4. J Virol. 2015. PMID: 25741006 Free PMC article.
-
Disifin (sodium tosylchloramide) and Toll-like receptors (TLRs): evolving importance in health and diseases.J Ind Microbiol Biotechnol. 2007 Dec;34(12):751-62. doi: 10.1007/s10295-007-0252-2. Epub 2007 Sep 5. J Ind Microbiol Biotechnol. 2007. PMID: 17876621 Review.
-
Avian influenza - The next pandemic?Can J Infect Dis Med Microbiol. 2004 Sep;15(5):252-4. doi: 10.1155/2004/121394. Can J Infect Dis Med Microbiol. 2004. PMID: 18159500 Free PMC article. No abstract available.
References
-
- Alexander D J, Lister S A, Johnson M J, Randall C J, Thomas P J. An outbreak of highly pathogenic avian influenza in turkeys in Great Britain in 1991. Vet Rec. 1993;132:535–536. - PubMed
-
- Alexander D J, Parsons G, Manvell R J. Experimental assessment of the pathogenicity of eight avian influenza A viruses of H5 subtype for chickens, turkeys, ducks and quail. Avian Pathol. 1986;15:647–662. - PubMed
-
- Almond J W. A single gene determines the host range of influenza virus. Nature (London) 1977;270:617–618. - PubMed
-
- Austin F J, Webster R G. Antigenic mapping of an avian H1 influenza virus haemagglutinin and interrelationships of H1 viruses from humans, pigs and birds. J Gen Virol. 1986;67:983–992. - PubMed
-
- Banks J, Speidel E, Alexander D J. Characterisation of an avian influenza A virus isolated from a human—is an intermediate host necessary for the emergence of pandemic influenza viruses? Arch Virol. 1998;143:781–787. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

