Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA
- PMID: 11139614
- PMCID: PMC29660
- DOI: 10.1093/nar/29.2.439
Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA
Abstract
DNMT2 is a human protein that displays strong sequence similarities to DNA (cytosine-5)-methyltransferases (m(5)C MTases) of both prokaryotes and eukaryotes. DNMT2 contains all 10 sequence motifs that are conserved among m(5)C MTases, including the consensus S:-adenosyl-L-methionine-binding motifs and the active site ProCys dipeptide. DNMT2 has close homologs in plants, insects and Schizosaccharomyces pombe, but no related sequence can be found in the genomes of Saccharomyces cerevisiae or Caenorhabditis elegans. The crystal structure of a deletion mutant of DNMT2 complexed with S-adenosyl-L-homocysteine (AdoHcy) has been determined at 1.8 A resolution. The structure of the large domain that contains the sequence motifs involved in catalysis is remarkably similar to that of M.HHAI, a confirmed bacterial m(5)C MTase, and the smaller target recognition domains of DNMT2 and M.HHAI are also closely related in overall structure. The small domain of DNMT2 contains three short helices that are not present in M.HHAI. DNMT2 binds AdoHcy in the same conformation as confirmed m(5)C MTases and, while DNMT2 shares all sequence and structural features with m(5)C MTases, it has failed to demonstrate detectable transmethylase activity. We show here that homologs of DNMT2, which are present in some organisms that are not known to methylate their genomes, contain a specific target-recognizing sequence motif including an invariant CysPheThr tripeptide. DNMT2 binds DNA to form a denaturant-resistant complex in vitro. While the biological function of DNMT2 is not yet known, the strong binding to DNA suggests that DNMT2 may mark specific sequences in the genome by binding to DNA through the specific target-recognizing motif.
Figures

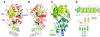
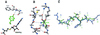
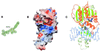
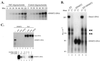
Similar articles
-
Structure analysis of Entamoeba histolytica DNMT2 (EhMeth).PLoS One. 2012;7(6):e38728. doi: 10.1371/journal.pone.0038728. Epub 2012 Jun 21. PLoS One. 2012. PMID: 22737219 Free PMC article.
-
Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-L-methionine.Cell. 1993 Jul 30;74(2):299-307. doi: 10.1016/0092-8674(93)90421-l. Cell. 1993. PMID: 8343957
-
M.phi 3TII: a new monospecific DNA (cytosine-C5) methyltransferase with pronounced amino acid sequence similarity to a family of adenine-N6-DNA-methyltransferases.Nucleic Acids Res. 1994 Oct 11;22(20):4066-72. doi: 10.1093/nar/22.20.4066. Nucleic Acids Res. 1994. Corrected and republished in: Nucleic Acids Res. 1994 Dec 11;22(24):5517-23. doi: 10.1093/nar/22.24.5517 PMID: 7937131 Free PMC article. Corrected and republished.
-
[Dnmt2 is the Most Evolutionary Conserved and Enigmatic Cytosine DNA Methyltransferase in Eukaryotes].Genetika. 2016 Mar;52(3):269-82. Genetika. 2016. PMID: 27281847 Review. Russian.
-
Mechanism and biological role of Dnmt2 in Nucleic Acid Methylation.RNA Biol. 2017 Sep 2;14(9):1108-1123. doi: 10.1080/15476286.2016.1191737. Epub 2016 May 27. RNA Biol. 2017. PMID: 27232191 Free PMC article. Review.
Cited by
-
The Drosophila cytosine-5 methyltransferase Dnmt2 is associated with the nuclear matrix and can access DNA during mitosis.PLoS One. 2008 Jan 9;3(1):e1414. doi: 10.1371/journal.pone.0001414. PLoS One. 2008. PMID: 18183295 Free PMC article.
-
Chemical Space Virtual Screening against Hard-to-Drug RNA Methyltransferases DNMT2 and NSUN6.Int J Mol Sci. 2023 Mar 24;24(7):6109. doi: 10.3390/ijms24076109. Int J Mol Sci. 2023. PMID: 37047081 Free PMC article.
-
Molecular Mechanisms Regulating Muscle Plasticity in Fish.Animals (Basel). 2020 Dec 30;11(1):61. doi: 10.3390/ani11010061. Animals (Basel). 2020. PMID: 33396941 Free PMC article. Review.
-
DNA methyltransferases in hematological malignancies.J Genet Genomics. 2020 Jul 20;47(7):361-372. doi: 10.1016/j.jgg.2020.04.006. Epub 2020 Jul 24. J Genet Genomics. 2020. PMID: 32994141 Free PMC article. Review.
-
Target recognition, RNA methylation activity and transcriptional regulation of the Dictyostelium discoideum Dnmt2-homologue (DnmA).Nucleic Acids Res. 2013 Oct;41(18):8615-27. doi: 10.1093/nar/gkt634. Epub 2013 Jul 22. Nucleic Acids Res. 2013. PMID: 23877245 Free PMC article.
References
-
- Bestor T.H. (2000) The DNA methyltransferases of mammals. Hum. Mol. Genet., 9, 2395–2402. - PubMed
-
- Lei H., Oh,S.P., Okano,M., Juttermann,R., Goss,K.A., Jaenisch,R. and Li,E. (1996) De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development, 122, 3195–3205. - PubMed
-
- Okano M., Bell,D.W., Haber,D.A. and Li,E. (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell, 99, 247–257. - PubMed
-
- Xu G.-L., Bestor,T.H., Bourc’his,D., Hsieh,C.-L., Tommerup,N., Bugge,M., Hulten,M., Qu,X., Russo,J.J. and Viegas-Pequignot,E. (1999) Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature, 402, 187–191. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

