p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks
- PMID: 11134068
- PMCID: PMC2150674
- DOI: 10.1083/jcb.151.7.1381
p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks
Abstract
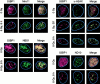
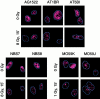
p53 binding protein 1 (53BP1), a protein proposed to function as a transcriptional coactivator of the p53 tumor suppressor, has BRCT domains with high homology to the Saccharomyces cerevisiae Rad9p DNA damage checkpoint protein. To examine whether 53BP1 has a role in the cellular response to DNA damage, we probed its intracellular localization by immunofluorescence. In untreated primary cells and U2OS osteosarcoma cells, 53BP1 exhibited diffuse nuclear staining; whereas, within 5-15 min after exposure to ionizing radiation (IR), 53BP1 localized at discreet nuclear foci. We propose that these foci represent sites of processing of DNA double-strand breaks (DSBs), because they were induced by IR and chemicals that cause DSBs, but not by ultraviolet light; their peak number approximated the number of DSBs induced by IR and decreased over time with kinetics that parallel the rate of DNA repair; and they colocalized with IR-induced Mre11/NBS and gamma-H2AX foci, which have been previously shown to localize at sites of DSBs. Formation of 53BP1 foci after irradiation was not dependent on ataxia-telangiectasia mutated (ATM), Nijmegen breakage syndrome (NBS1), or wild-type p53. Thus, the fast kinetics of 53BP1 focus formation after irradiation and the lack of dependency on ATM and NBS1 suggest that 53BP1 functions early in the cellular response to DNA DSBs.
Figures





Similar articles
-
Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage.Mol Cell Biol. 2001 Mar;21(5):1719-29. doi: 10.1128/MCB.21.5.1719-1729.2001. Mol Cell Biol. 2001. PMID: 11238909 Free PMC article.
-
53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair.Nat Cell Biol. 2010 Feb;12(2):177-84. doi: 10.1038/ncb2017. Epub 2010 Jan 17. Nat Cell Biol. 2010. PMID: 20081839
-
Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways.J Cell Biol. 2001 Apr 30;153(3):613-20. doi: 10.1083/jcb.153.3.613. J Cell Biol. 2001. PMID: 11331310 Free PMC article.
-
ATM signaling and 53BP1.Radiother Oncol. 2005 Aug;76(2):119-22. doi: 10.1016/j.radonc.2005.06.026. Radiother Oncol. 2005. PMID: 16024119 Review.
-
The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax.DNA Repair (Amst). 2010 Dec 10;9(12):1273-82. doi: 10.1016/j.dnarep.2010.09.013. Epub 2010 Oct 30. DNA Repair (Amst). 2010. PMID: 21036673 Review.
Cited by
-
Annexin A2: the importance of being redox sensitive.Int J Mol Sci. 2013 Feb 7;14(2):3568-94. doi: 10.3390/ijms14023568. Int J Mol Sci. 2013. PMID: 23434659 Free PMC article.
-
DNA polymerase ζ is a major determinant of resistance to platinum-based chemotherapeutic agents.Mol Pharmacol. 2012 Jun;81(6):778-87. doi: 10.1124/mol.111.076828. Epub 2012 Mar 2. Mol Pharmacol. 2012. PMID: 22387291 Free PMC article.
-
WRN helicase regulates the ATR-CHK1-induced S-phase checkpoint pathway in response to topoisomerase-I-DNA covalent complexes.J Cell Sci. 2011 Dec 1;124(Pt 23):3967-79. doi: 10.1242/jcs.081372. Epub 2011 Dec 8. J Cell Sci. 2011. PMID: 22159421 Free PMC article.
-
The Focinator - a new open-source tool for high-throughput foci evaluation of DNA damage.Radiat Oncol. 2015 Aug 4;10:163. doi: 10.1186/s13014-015-0453-1. Radiat Oncol. 2015. PMID: 26238507 Free PMC article.
-
DNA Damage Foci on Metaphase Chromosomes.Methods Mol Biol. 2023;2519:93-98. doi: 10.1007/978-1-0716-2433-3_10. Methods Mol Biol. 2023. PMID: 36066713
References
-
- Banin S., Moyal L., Shieh S., Taya Y., Anderson C.W., Chessa L., Smorodinsky N.I., Prives C., Reiss Y., Shiloh Y., Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. - PubMed
-
- Bao S., Shen X., Shen K., Liu Y., Wang X.F. The mammalian Rad24 homologous to yeast Saccharomyces cerevisiae Rad24 and Schizosaccharomyces pombe Rad17 is involved in DNA damage checkpoint. Cell Growth Differ. 1998;9:961–967. - PubMed
-
- Blasina A., de Weyer I.V., Laus M.C., Luyten W.H., Parker A.E., McGowan C.H. A human homologue of the checkpoint kinase Cds1 directly inhibits Cdc25 phosphatase Curr. Biol. 9 1999. 1 10a - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

