cis-acting sequences required for coronavirus infectious bronchitis virus defective-RNA replication and packaging
- PMID: 11119581
- PMCID: PMC113905
- DOI: 10.1128/JVI.75.1.125-133.2001
cis-acting sequences required for coronavirus infectious bronchitis virus defective-RNA replication and packaging
Abstract
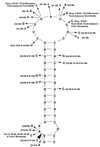
The parts of the RNA genome of infectious bronchitis virus (IBV) required for replication and packaging of the RNA were investigated using deletion mutagenesis of a defective RNA (D-RNA) CD-61 (6.1 kb) containing a chloramphenicol acetyltransferase reporter gene. A D-RNA with the first 544, but not as few as 338, nucleotides (nt) of the 5' terminus was replicated; the 5' untranslated region (UTR) comprises 528 nt. Region I of the 3' UTR, adjacent to the nucleocapsid protein gene, comprised 212 nt and could be removed without impairment of replication or packaging of D-RNAs. A D-RNA with the final 338 nt, including the 293 nt in the highly conserved region II of the 3' UTR, was replicated. Thus, the 5'-terminal 544 nt and 3'-terminal 338 nt contained the necessary signals for RNA replication. Phylogenetic analysis of 19 strains of IBV and 3 strains of turkey coronavirus predicted a conserved stem-loop structure at the 5' end of region II of the 3' UTR. Removal of the predicted stem-loop structure abolished replication of the D-RNAs. D-RNAs in which replicase gene 1b-derived sequences had been removed or replaced with all the downstream genes were replicated well but were rescued poorly, suggesting inefficient packaging. However, no specific part of the 1b gene was required for efficient packaging.
Figures



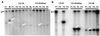

Similar articles
-
Leader switching occurs during the rescue of defective RNAs by heterologous strains of the coronavirus infectious bronchitis virus.J Gen Virol. 2000 Mar;81(Pt 3):791-801. doi: 10.1099/0022-1317-81-3-791. J Gen Virol. 2000. PMID: 10675417
-
Replication and packaging of coronavirus infectious bronchitis virus defective RNAs lacking a long open reading frame.J Virol. 1996 Dec;70(12):8660-8. doi: 10.1128/JVI.70.12.8660-8668.1996. J Virol. 1996. PMID: 8970992 Free PMC article.
-
Utilizing fowlpox virus recombinants to generate defective RNAs of the coronavirus infectious bronchitis virus.J Gen Virol. 2000 Dec;81(Pt 12):2855-2865. doi: 10.1099/0022-1317-81-12-2855. J Gen Virol. 2000. PMID: 11086116
-
Sequence elements involved in the rescue of IBV defective RNA CD-91.Adv Exp Med Biol. 1998;440:253-7. doi: 10.1007/978-1-4615-5331-1_32. Adv Exp Med Biol. 1998. PMID: 9782289
-
Translation and replication of FMDV RNA.Curr Top Microbiol Immunol. 2005;288:43-70. doi: 10.1007/3-540-27109-0_3. Curr Top Microbiol Immunol. 2005. PMID: 15648174 Review.
Cited by
-
Enhanced accumulation of coronavirus defective interfering RNA from expressed negative-strand transcripts by coexpressed positive-strand RNA transcripts.Virology. 2001 Sep 1;287(2):286-300. doi: 10.1006/viro.2001.1047. Virology. 2001. PMID: 11531407 Free PMC article.
-
Stem-loop III in the 5' untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication.J Virol. 2003 Jun;77(12):6720-30. doi: 10.1128/jvi.77.12.6720-6730.2003. J Virol. 2003. PMID: 12767992 Free PMC article.
-
Coronavirus cis-Acting RNA Elements.Adv Virus Res. 2016;96:127-163. doi: 10.1016/bs.aivir.2016.08.007. Epub 2016 Sep 6. Adv Virus Res. 2016. PMID: 27712622 Free PMC article. Review.
-
Coronavirus genomic RNA packaging.Virology. 2019 Nov;537:198-207. doi: 10.1016/j.virol.2019.08.031. Epub 2019 Aug 30. Virology. 2019. PMID: 31505321 Free PMC article. Review.
-
The molecular biology of coronaviruses.Adv Virus Res. 2006;66:193-292. doi: 10.1016/S0065-3527(06)66005-3. Adv Virus Res. 2006. PMID: 16877062 Free PMC article. Review.
References
-
- Boursnell M E G, Binns M M, Foulds I J, Brown T D K. Sequences of the nucleocapsid genes from two strains of avian infectious bronchitis virus. J Gen Virol. 1985;66:573–580. - PubMed
-
- Brian D A, Chang R-Y, Hofmann M A, Sethna P B. Role of subgenomic minus-strand RNA in coronavirus replication. Arch Virol. 1994;9(Suppl.):173–180. - PubMed
-
- Cavanagh D, Brian D A, Brinton M A, Enjuanes L, Holmes K V, Horzinek M C, Lai M M C, Laude H, Plagemann P G W, Siddell S G, Spaan W, Taguchi F, Talbot P J. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

