Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4)
- PMID: 11113196
- PMCID: PMC88795
- DOI: 10.1128/MCB.21.1.209-223.2001
Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4)
Abstract
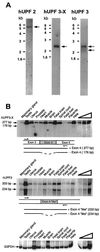
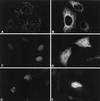
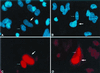
Nonsense-mediated mRNA decay (NMD), also called mRNA surveillance, is an important pathway used by all organisms that have been tested to degrade mRNAs that prematurely terminate translation and, as a consequence, eliminate the production of aberrant proteins that could be potentially harmful. In mammalian cells, NMD appears to involve splicing-dependent alterations to mRNA as well as ribosome-associated components of the translational apparatus. To date, human (h) Upf1 protein (p) (hUpf1p), a group 1 RNA helicase named after its Saccharomyces cerevisiae orthologue that functions in both translation termination and NMD, has been the only factor shown to be required for NMD in mammalian cells. Here, we describe human orthologues to S. cerevisiae Upf2p and S. cerevisiae Upf3p (Caenorhabditis elegans SMG-4) based on limited amino acid similarities. The existence of these orthologues provides evidence for a higher degree of evolutionary conservation of NMD than previously appreciated. Interestingly, human orthologues to S. cerevisiae Upf3p (C. elegans SMG-4) derive from two genes, one of which is X-linked and both of which generate multiple isoforms due to alternative pre-mRNA splicing. We demonstrate using immunoprecipitations of epitope-tagged proteins transiently produced in HeLa cells that hUpf2p interacts with hUpf1p, hUpf3p-X, and hUpf3p, and we define the domains required for the interactions. Furthermore, we find by using indirect immunofluorescence that hUpf1p is detected only in the cytoplasm, hUpf2p is detected primarily in the cytoplasm, and hUpf3p-X localizes primarily to nuclei. The finding that hUpf3p-X is a shuttling protein provides additional indication that NMD has both nuclear and cytoplasmic components.
Figures







Similar articles
-
Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes.Mol Cell Biol. 2000 Dec;20(23):8944-57. doi: 10.1128/MCB.20.23.8944-8957.2000. Mol Cell Biol. 2000. PMID: 11073994 Free PMC article.
-
A highly conserved region essential for NMD in the Upf2 N-terminal domain.J Mol Biol. 2014 Nov 11;426(22):3689-3702. doi: 10.1016/j.jmb.2014.09.015. Epub 2014 Sep 30. J Mol Biol. 2014. PMID: 25277656 Free PMC article.
-
Caenorhabditis elegans SMG-2 selectively marks mRNAs containing premature translation termination codons.Mol Cell Biol. 2007 Aug;27(16):5630-8. doi: 10.1128/MCB.00410-07. Epub 2007 Jun 11. Mol Cell Biol. 2007. PMID: 17562857 Free PMC article.
-
NMD monitors translational fidelity 24/7.Curr Genet. 2017 Dec;63(6):1007-1010. doi: 10.1007/s00294-017-0709-4. Epub 2017 May 23. Curr Genet. 2017. PMID: 28536849 Free PMC article. Review.
-
Upf proteins: highly conserved factors involved in nonsense mRNA mediated decay.Mol Biol Rep. 2018 Feb;45(1):39-55. doi: 10.1007/s11033-017-4139-7. Epub 2017 Dec 27. Mol Biol Rep. 2018. PMID: 29282598 Review.
Cited by
-
Nonsense-mediated mRNA decay (NMD) in animal embryogenesis: to die or not to die, that is the question.Curr Opin Genet Dev. 2011 Aug;21(4):422-30. doi: 10.1016/j.gde.2011.03.008. Curr Opin Genet Dev. 2011. PMID: 21550797 Free PMC article. Review.
-
Mechanism, factors, and physiological role of nonsense-mediated mRNA decay.Cell Mol Life Sci. 2015 Dec;72(23):4523-44. doi: 10.1007/s00018-015-2017-9. Epub 2015 Aug 18. Cell Mol Life Sci. 2015. PMID: 26283621 Free PMC article. Review.
-
Phospho-dependent and phospho-independent interactions of the helicase UPF1 with the NMD factors SMG5-SMG7 and SMG6.Nucleic Acids Res. 2014 Aug;42(14):9447-60. doi: 10.1093/nar/gku578. Epub 2014 Jul 10. Nucleic Acids Res. 2014. PMID: 25013172 Free PMC article.
-
The Antagonistic Gene Paralogs Upf3a and Upf3b Govern Nonsense-Mediated RNA Decay.Cell. 2016 Apr 7;165(2):382-95. doi: 10.1016/j.cell.2016.02.046. Epub 2016 Mar 31. Cell. 2016. PMID: 27040500 Free PMC article.
-
A protein interaction framework for human mRNA degradation.Genome Res. 2004 Jul;14(7):1315-23. doi: 10.1101/gr.2122004. Genome Res. 2004. PMID: 15231747 Free PMC article.
References
-
- Atkin A L, Schenkman L R, Eastham M, Dahlseid J N, Lelivelt M J, Culbertson M R. Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J Biol Chem. 1997;272:22163–22172. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
