Analysis of the steroid receptor coactivator 1 (SRC1)-CREB binding protein interaction interface and its importance for the function of SRC1
- PMID: 11113179
- PMCID: PMC86566
- DOI: 10.1128/MCB.21.1.39-50.2001
Analysis of the steroid receptor coactivator 1 (SRC1)-CREB binding protein interaction interface and its importance for the function of SRC1
Abstract
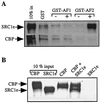
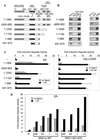
The transcriptional activity of nuclear receptors is mediated by coactivator proteins, including steroid receptor coactivator 1 (SRC1) and its homologues and the general coactivators CREB binding protein (CBP) and p300. SRC1 contains an activation domain (AD1) which functions via recruitment of CBP and and p300. In this study, we have used yeast two-hybrid and in vitro interaction-peptide inhibition experiments to map the AD1 domain of SRC1 to a 35-residue sequence potentially containing two alpha-helices. We also define a 72-amino-acid sequence in CBP necessary for SRC1 binding, designated the SRC1 interaction domain (SID). We show that in contrast to SRC1, direct binding of CBP to the estrogen receptor is weak, suggesting that SRC1 functions primarily as an adaptor to recruit CBP and p300. In support of this, we show that the ability of SRC1 to enhance ligand-dependent nuclear receptor activity in transiently transfected cells is dependent upon the integrity of the AD1 region. In contrast, the putative histone acetyltransferase domain, the Per-Arnt-Sim basic helix-loop-helix domain, the glutamine-rich domain, and AD2 can each be removed without loss of ligand-induced activity. Remarkably, a construct corresponding to residues 631 to 970, which contains only the LXXLL motifs and the AD1 region of SRC1, retained strong coactivator activity in our assays.
Figures




Similar articles
-
A Conserved alpha-helical motif mediates the binding of diverse nuclear proteins to the SRC1 interaction domain of CBP.J Biol Chem. 2004 Apr 2;279(14):14055-64. doi: 10.1074/jbc.M310188200. Epub 2004 Jan 13. J Biol Chem. 2004. PMID: 14722092
-
Core LXXLL motif sequences in CREB-binding protein, SRC1, and RIP140 define affinity and selectivity for steroid and retinoid receptors.J Biol Chem. 2001 Mar 2;276(9):6695-702. doi: 10.1074/jbc.M009404200. Epub 2000 Nov 14. J Biol Chem. 2001. PMID: 11078741
-
Transcriptional activation by estrogen receptor (ERalpha) and steroid receptor coactivator (SRC1) involves distinct mechanisms in yeast and mammalian cells.J Mol Endocrinol. 2003 Jun;30(3):411-22. doi: 10.1677/jme.0.0300411. J Mol Endocrinol. 2003. PMID: 12790809
-
Mechanisms of androgen receptor signalling via steroid receptor coactivator-1 in prostate.Endocr Relat Cancer. 2004 Mar;11(1):117-30. doi: 10.1677/erc.0.0110117. Endocr Relat Cancer. 2004. PMID: 15027889
-
Structural diversity in p160/CREB-binding protein coactivator complexes.J Biol Chem. 2006 May 26;281(21):14787-95. doi: 10.1074/jbc.M600237200. Epub 2006 Mar 15. J Biol Chem. 2006. PMID: 16540468
Cited by
-
GAS, a new glutamate-rich protein, interacts differentially with SRCs and is involved in oestrogen receptor function.EMBO Rep. 2009 Jan;10(1):51-7. doi: 10.1038/embor.2008.223. Epub 2008 Nov 28. EMBO Rep. 2009. PMID: 19039327 Free PMC article.
-
P160/SRC/NCoA coactivators form complexes via specific interaction of their PAS-B domain with the CID/AD1 domain.Nucleic Acids Res. 2008 Apr;36(6):1847-60. doi: 10.1093/nar/gkn029. Epub 2008 Feb 11. Nucleic Acids Res. 2008. PMID: 18267973 Free PMC article.
-
Dynamic assembly and activation of estrogen receptor α enhancers through coregulator switching.Genes Dev. 2017 Aug 1;31(15):1535-1548. doi: 10.1101/gad.302182.117. Epub 2017 Sep 8. Genes Dev. 2017. PMID: 28887413 Free PMC article.
-
MOZ-TIF2 inhibits transcription by nuclear receptors and p53 by impairment of CBP function.Mol Cell Biol. 2005 Feb;25(3):988-1002. doi: 10.1128/MCB.25.3.988-1002.2005. Mol Cell Biol. 2005. PMID: 15657427 Free PMC article.
-
Intestinal remodeling during Xenopus metamorphosis as a model for studying thyroid hormone signaling and adult organogenesis.Mol Cell Endocrinol. 2024 May 15;586:112193. doi: 10.1016/j.mce.2024.112193. Epub 2024 Feb 22. Mol Cell Endocrinol. 2024. PMID: 38401883
References
-
- Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X-Y, Sauter G, Kallioniemi O-P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. - PubMed
-
- Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
