Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose
- PMID: 11106374
- PMCID: PMC18891
- DOI: 10.1073/pnas.250422697
Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose
Abstract
Conflicting reports have suggested that the silent information regulator 2 (SIR2) protein family employs NAD(+) to ADP-ribosylate histones [Tanny, J. C., Dowd, G. J., Huang, J., Hilz, H. & Moazed, D. (1999) Cell 99, 735-745; Frye, R. A. (1999) Biochem. Biophys. Res. Commun. 260, 273-279], deacetylate histones [Landry, J., Sutton, A., Tafrov, S. T., Heller, R. C., Stebbins, J., Pillus, L. & Sternglanz, R. (2000) Proc. Natl. Acad. Sci. USA 97, 5807-5811; Smith, J. S., Brachmann, C. B., Celic, I., Kenna, M. A., Muhammad, S., Starai, V. J., Avalos, J. L., Escalante-Semerena, J. C., Grubmeyer, C., Wolberger, C. & Boeke, J. D. (2000) Proc. Natl. Acad. Sci. USA 97, 6658-6663], or both [Imai, S., Armstrong, C. M., Kaeberlein, M. & Guarente, L. (2000) Nature (London) 403, 795-800]. Uncovering the true enzymatic function of SIR2 is critical to the basic understanding of its cellular function. Therefore, we set out to authenticate the reaction products and to determine the intrinsic catalytic mechanism. We provide direct evidence that the efficient histone/protein deacetylase reaction is tightly coupled to the formation of a previously unidentified acetyl-ADP-ribose product (1-O-acetyl-ADP ribose). One molecule of NAD(+) and one molecule of acetyl-lysine are readily catalyzed to one molecule of deacetylated lysine, nicotinamide, and 1-O-acetyl-ADP-ribose. A unique reaction mechanism involving the attack of enzyme-bound acetate or the direct attack of acetyl-lysine on an oxocarbenium ADP-ribose intermediate is proposed. We suggest that the reported histone/protein ADP-ribosyltransferase activity is a low-efficiency side reaction that can be explained through the partial uncoupling of the intrinsic deacetylation and acetate transfer to ADP-ribose.
Figures
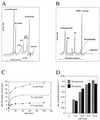
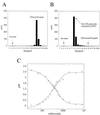
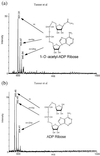
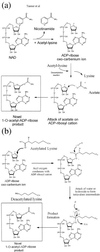
Comment in
-
The Sir2 protein family: A novel deacetylase for gene silencing and more.Proc Natl Acad Sci U S A. 2000 Dec 19;97(26):14030-2. doi: 10.1073/pnas.011506198. Proc Natl Acad Sci U S A. 2000. PMID: 11114164 Free PMC article. Review. No abstract available.
Similar articles
-
Structural identification of 2'- and 3'-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of beta -NAD+-dependent histone/protein deacetylases.J Biol Chem. 2002 May 24;277(21):18535-44. doi: 10.1074/jbc.M200671200. Epub 2002 Mar 13. J Biol Chem. 2002. PMID: 11893743
-
Conserved enzymatic production and biological effect of O-acetyl-ADP-ribose by silent information regulator 2-like NAD+-dependent deacetylases.J Biol Chem. 2002 Apr 12;277(15):12632-41. doi: 10.1074/jbc.M111830200. Epub 2002 Jan 25. J Biol Chem. 2002. PMID: 11812793
-
Role of NAD(+) in the deacetylase activity of the SIR2-like proteins.Biochem Biophys Res Commun. 2000 Nov 30;278(3):685-90. doi: 10.1006/bbrc.2000.3854. Biochem Biophys Res Commun. 2000. PMID: 11095969
-
SIR2: the biochemical mechanism of NAD(+)-dependent protein deacetylation and ADP-ribosyl enzyme intermediates.Curr Med Chem. 2004 Apr;11(7):807-26. doi: 10.2174/0929867043455675. Curr Med Chem. 2004. PMID: 15078167 Review.
-
Enzymatic activities of Sir2 and chromatin silencing.Curr Opin Cell Biol. 2001 Apr;13(2):232-8. doi: 10.1016/s0955-0674(00)00202-7. Curr Opin Cell Biol. 2001. PMID: 11248558 Review.
Cited by
-
Structural basis for allosteric, substrate-dependent stimulation of SIRT1 activity by resveratrol.Genes Dev. 2015 Jun 15;29(12):1316-25. doi: 10.1101/gad.265462.115. Genes Dev. 2015. PMID: 26109052 Free PMC article.
-
The role of multiple marks in epigenetic silencing and the emergence of a stable bivalent chromatin state.PLoS Comput Biol. 2013;9(7):e1003121. doi: 10.1371/journal.pcbi.1003121. Epub 2013 Jul 18. PLoS Comput Biol. 2013. PMID: 23874171 Free PMC article.
-
Histone deacetylase inhibitors (HDACIs): multitargeted anticancer agents.Biologics. 2013;7:47-60. doi: 10.2147/BTT.S29965. Epub 2013 Feb 25. Biologics. 2013. PMID: 23459471 Free PMC article.
-
Early steps in the biosynthesis of NAD in Arabidopsis start with aspartate and occur in the plastid.Plant Physiol. 2006 Jul;141(3):851-7. doi: 10.1104/pp.106.081091. Epub 2006 May 12. Plant Physiol. 2006. PMID: 16698895 Free PMC article.
-
Nicotinamide riboside kinase structures reveal new pathways to NAD+.PLoS Biol. 2007 Oct 2;5(10):e263. doi: 10.1371/journal.pbio.0050263. PLoS Biol. 2007. PMID: 17914902 Free PMC article.
References
-
- Frye R A. Biochem Biophys Res Commun. 2000;273:793–798. - PubMed
-
- Loo S, Rine J. Annu Rev Cell Dev Biol. 1995;11:519–548. - PubMed
-
- Gottlieb S, Esposito R E. Cell. 1989;56:771–776. - PubMed
-
- Guarente L. Nat Genet. 1999;23:281–285. - PubMed
-
- Brachmann C B, Sherman J M, Devine S E, Cameron E E, Pillus L, Boeke J D. Genes Dev. 1995;9:2888–2902. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

