Preferential relaxation of positively supercoiled DNA by E. coli topoisomerase IV in single-molecule and ensemble measurements
- PMID: 11090135
- PMCID: PMC317058
- DOI: 10.1101/gad.838900
Preferential relaxation of positively supercoiled DNA by E. coli topoisomerase IV in single-molecule and ensemble measurements
Abstract
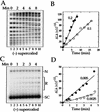
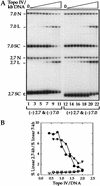
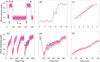
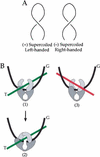
We show that positively supercoiled [(+) SC] DNA is the preferred substrate for Escherichia coli topoisomerase IV (topo IV). We measured topo IV relaxation of (-) and (+) supercoils in real time on single, tethered DNA molecules to complement ensemble experiments. We find that the preference for (+) SC DNA is complete at low enzyme concentration. Otherwise, topo IV relaxed (+) supercoils at a 20-fold faster rate than (-) supercoils, due primarily to about a 10-fold increase in processivity with (+) SC DNA. The preferential cleavage of (+) SC DNA in a competition experiment showed that substrate discrimination can take place prior to strand passage in the presence or absence of ATP. We propose that topo IV discriminates between (-) and (+) supercoiled DNA by recognition of the geometry of (+) SC DNA. Our results explain how topo IV can rapidly remove (+) supercoils to support DNA replication without relaxing the essential (-) supercoils of the chromosome. They also show that the rate of supercoil relaxation by topo IV is several orders of magnitude faster than hitherto appreciated, so that a single enzyme may suffice at each replication fork.
Figures


Similar articles
-
Chirality sensing by Escherichia coli topoisomerase IV and the mechanism of type II topoisomerases.Proc Natl Acad Sci U S A. 2003 Jul 22;100(15):8654-9. doi: 10.1073/pnas.1133178100. Epub 2003 Jul 11. Proc Natl Acad Sci U S A. 2003. PMID: 12857958 Free PMC article.
-
Chiral discrimination and writhe-dependent relaxation mechanism of human topoisomerase IIα.J Biol Chem. 2013 May 10;288(19):13695-703. doi: 10.1074/jbc.M112.444745. Epub 2013 Mar 18. J Biol Chem. 2013. PMID: 23508957 Free PMC article.
-
Activities of gyrase and topoisomerase IV on positively supercoiled DNA.Nucleic Acids Res. 2017 Sep 19;45(16):9611-9624. doi: 10.1093/nar/gkx649. Nucleic Acids Res. 2017. PMID: 28934496 Free PMC article.
-
[DNA supercoiling and topoisomerases in Escherichia coli].Rev Latinoam Microbiol. 1995 Jul-Sep;37(3):291-304. Rev Latinoam Microbiol. 1995. PMID: 8850348 Review. Spanish.
-
DNA supercoiling and relaxation by ATP-dependent DNA topoisomerases.Philos Trans R Soc Lond B Biol Sci. 1992 Apr 29;336(1276):83-91. doi: 10.1098/rstb.1992.0047. Philos Trans R Soc Lond B Biol Sci. 1992. PMID: 1351300 Review.
Cited by
-
Topological stress is responsible for the detrimental outcomes of head-on replication-transcription conflicts.Cell Rep. 2021 Mar 2;34(9):108797. doi: 10.1016/j.celrep.2021.108797. Cell Rep. 2021. PMID: 33657379 Free PMC article.
-
The MukB-ParC interaction affects the intramolecular, not intermolecular, activities of topoisomerase IV.J Biol Chem. 2013 Mar 15;288(11):7653-7661. doi: 10.1074/jbc.M112.418087. Epub 2013 Jan 24. J Biol Chem. 2013. PMID: 23349462 Free PMC article.
-
Recognition of DNA Supercoil Handedness during Catenation Catalyzed by Type II Topoisomerases.Biochemistry. 2022 Oct 4;61(19):2148-2158. doi: 10.1021/acs.biochem.2c00370. Epub 2022 Sep 19. Biochemistry. 2022. PMID: 36122251 Free PMC article.
-
Mechanisms of chiral discrimination by topoisomerase IV.Proc Natl Acad Sci U S A. 2009 Apr 28;106(17):6986-91. doi: 10.1073/pnas.0900574106. Epub 2009 Apr 9. Proc Natl Acad Sci U S A. 2009. PMID: 19359479 Free PMC article.
-
The African Swine Fever Virus (ASFV) Topoisomerase II as a Target for Viral Prevention and Control.Vaccines (Basel). 2020 Jun 17;8(2):312. doi: 10.3390/vaccines8020312. Vaccines (Basel). 2020. PMID: 32560397 Free PMC article. Review.
References
-
- Benjamin HW, Cozzarelli NR. Geometric arrangements of Tn3 resolvase sites. J Biol Chem. 1990;265:6441–6447. - PubMed
-
- Berger JM, Gamblin SJ, Harrison SC, Wang JC. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. - PubMed
-
- Bird R, Lark K. Chromosome replication in Escherichia coli 15T– at different growth rates: Rate of replication of the chromosome and the rate of formation of small pieces. J Mol Biol. 1970;49:343–366. - PubMed
-
- Boocock MR, Brown JL, Sherratt DJ. Topological specificity in Tn3 resolvase catalysis. In: McMacken R, Kelly TJ, editors. DNA replication and recombination. Vol. 47. New York: Alan R. Liss; 1987. pp. 703–718.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
