Functional interaction between nucleosome assembly proteins and p300/CREB-binding protein family coactivators
- PMID: 11073993
- PMCID: PMC86548
- DOI: 10.1128/MCB.20.23.8933-8943.2000
Functional interaction between nucleosome assembly proteins and p300/CREB-binding protein family coactivators
Abstract
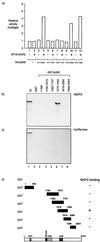
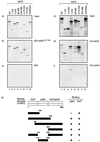
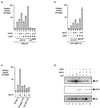
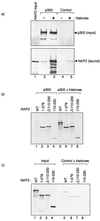
The p300/CREB-binding protein (CBP) family of proteins consists of coactivators that influence the activity of a wide variety of transcription factors. Although the mechanisms that allow p300/CBP proteins to achieve transcriptional control are not clear, it is believed that the regulation of chromatin is an important aspect of the process. Here, we describe a new level of p300-dependent control mediated through the functional interaction between p300/CBP and members of the family of nucleosome assembly proteins (NAP), which includes NAP1, NAP2, and TAF1. We find that NAP proteins, which have previously been implicated in the regulation of transcription factor binding to chromatin, augment the activity of different p300 targets, including p53 and E2F, through a process that is likely to involve the physical interaction between p300 and NAP. NAP proteins can form oligomers, and the results show that NAP proteins can bind to both core histones and p300 coactivator proteins, perhaps in a multicomponent ternary complex. We also provide data in support of the idea that histones can influence the interaction between p300 and NAP protein. These results argue that NAP is a functionally important component of the p300 coactivator complex and suggest that NAP may serve as a point of integration between transcriptional coactivators and chromatin.
Figures


Similar articles
-
Dual roles of p300 in chromatin assembly and transcriptional activation in cooperation with nucleosome assembly protein 1 in vitro.Mol Cell Biol. 2002 May;22(9):2974-83. doi: 10.1128/MCB.22.9.2974-2983.2002. Mol Cell Biol. 2002. PMID: 11940655 Free PMC article.
-
The coactivators CBP/p300 and the histone chaperone NAP1 promote transcription-independent nucleosome eviction at the HTLV-1 promoter.Proc Natl Acad Sci U S A. 2008 Jun 10;105(23):7959-63. doi: 10.1073/pnas.0800534105. Epub 2008 Jun 3. Proc Natl Acad Sci U S A. 2008. PMID: 18523016 Free PMC article.
-
p300-mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone.Genes Dev. 2000 Aug 1;14(15):1899-907. Genes Dev. 2000. PMID: 10921904 Free PMC article.
-
p300/CBP/p53 interaction and regulation of the p53 response.Eur J Biochem. 2001 May;268(10):2773-8. doi: 10.1046/j.1432-1327.2001.02226.x. Eur J Biochem. 2001. PMID: 11358491 Review.
-
CBP and p300: HATs for different occasions.Biochem Pharmacol. 2004 Sep 15;68(6):1145-55. doi: 10.1016/j.bcp.2004.03.045. Biochem Pharmacol. 2004. PMID: 15313412 Review.
Cited by
-
Epstein-Barr virus protein can upregulate cyclo-oxygenase-2 expression through association with the suppressor of metastasis Nm23-H1.J Virol. 2006 Feb;80(3):1321-31. doi: 10.1128/JVI.80.3.1321-1331.2006. J Virol. 2006. PMID: 16415009 Free PMC article.
-
Development of histone deacetylase inhibitors as therapeutics for neurological disease.Future Neurol. 2009 Nov 1;4(6):775-784. doi: 10.2217/fnl.09.55. Future Neurol. 2009. PMID: 20177429 Free PMC article.
-
Chromatin-specific remodeling by HMGB1 and linker histone H1 silences proinflammatory genes during endotoxin tolerance.Mol Cell Biol. 2009 Apr;29(7):1959-71. doi: 10.1128/MCB.01862-08. Epub 2009 Jan 21. Mol Cell Biol. 2009. PMID: 19158276 Free PMC article.
-
Testis specific Y-like 5: gene expression, methylation and implications for drug sensitivity in prostate carcinoma.BMC Cancer. 2017 Feb 24;17(1):158. doi: 10.1186/s12885-017-3134-7. BMC Cancer. 2017. PMID: 28235398 Free PMC article.
-
NSBP-1 mediates the effects of cholesterol on insulin/IGF-1 signaling in Caenorhabditis elegans.Cell Mol Life Sci. 2013 May;70(9):1623-36. doi: 10.1007/s00018-012-1221-0. Epub 2012 Dec 20. Cell Mol Life Sci. 2013. PMID: 23255046 Free PMC article.
References
-
- Adams C R, Kamakaka R T. Chromatin assembly: biochemical identities and genetic redundancy. Curr Opin Genet Dev. 1999;9:185–190. - PubMed
-
- Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. - PubMed
-
- Armstrong J A, Emerson B M. Transcription of chromatin: these are complex times. Curr Opin Genet Dev. 1998;8:165–172. - PubMed
-
- Avantaggiati M L, Ogryzko V V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. - PubMed
-
- Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
