Retroviral cDNA integration: stimulation by HMG I family proteins
- PMID: 11069991
- PMCID: PMC113176
- DOI: 10.1128/jvi.74.23.10965-10974.2000
Retroviral cDNA integration: stimulation by HMG I family proteins
Abstract
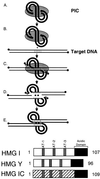
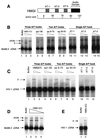
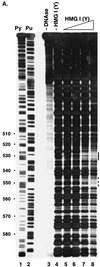
To replicate, a retrovirus must synthesize a cDNA copy of the viral RNA genome and integrate that cDNA into a chromosome of the host. We have investigated the role of a host cell cofactor, HMG I(Y) protein, in integration of human immunodeficiency virus type 1 (HIV-1) and Moloney murine leukemia virus (MoMLV) cDNA. Previously we reported that HMG I(Y) cofractionates with HIV-1 preintegration complexes (PICs) isolated from freshly infected cells. PICs depleted of required components by treatment with high concentrations of salt could be reconstituted by addition of purified HMG I(Y) in vitro. Here we report studies using immunoprecipitation that indicate that HMG I(Y) is associated with MoMLV preintegration complexes. In mechanistic studies, we show for both HIV-1 and MoMLV that each HMG I(Y) monomer must contain multiple DNA binding domains to stimulate integration by HMG I(Y)-depleted PICs. We also find that HMG I(Y) can condense model HIV-1 or MoMLV cDNA in vitro as measured by stimulation of intermolecular ligation. This reaction, like reconstitution of integration, depends on the presence of multiple DNA binding domains in each HMG I(Y) monomer. These data suggest that binding of multivalent HMG I(Y) monomers to multiple cDNA sites compacts retroviral cDNA, thereby promoting formation of active integrase-cDNA complexes.
Figures
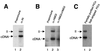


Similar articles
-
Modulation of activity of Moloney murine leukemia virus preintegration complexes by host factors in vitro.J Virol. 1998 Mar;72(3):2125-31. doi: 10.1128/JVI.72.3.2125-2131.1998. J Virol. 1998. PMID: 9499068 Free PMC article.
-
HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro.Cell. 1997 Feb 21;88(4):483-92. doi: 10.1016/s0092-8674(00)81888-7. Cell. 1997. PMID: 9038339
-
Transcription factor YY1 interacts with retroviral integrases and facilitates integration of moloney murine leukemia virus cDNA into the host chromosomes.J Virol. 2010 Aug;84(16):8250-61. doi: 10.1128/JVI.02681-09. Epub 2010 Jun 2. J Virol. 2010. PMID: 20519390 Free PMC article.
-
Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition.J Virol. 1997 Jul;71(7):5382-90. doi: 10.1128/JVI.71.7.5382-5390.1997. J Virol. 1997. PMID: 9188609 Free PMC article.
-
[Host factors that regulate the intercellular dynamics of HIV-1 genome during the early phase of infection].Uirusu. 2006 Jun;56(1):41-50. doi: 10.2222/jsv.56.41. Uirusu. 2006. PMID: 17038811 Review. Japanese.
Cited by
-
Specific inhibition of human immunodeficiency virus type 1 (HIV-1) integration in cell culture: putative inhibitors of HIV-1 integrase.Antimicrob Agents Chemother. 2001 Sep;45(9):2510-6. doi: 10.1128/AAC.45.9.2510-2516.2001. Antimicrob Agents Chemother. 2001. PMID: 11502522 Free PMC article.
-
Kinetics of human immunodeficiency virus type 1 (HIV) DNA integration in acutely infected cells as determined using a novel assay for detection of integrated HIV DNA.J Virol. 2001 Nov;75(22):11253-60. doi: 10.1128/JVI.75.22.11253-11260.2001. J Virol. 2001. PMID: 11602768 Free PMC article.
-
Cofactors for human immunodeficiency virus type 1 cDNA integration in vitro.J Virol. 2003 Jan;77(2):1598-603. doi: 10.1128/jvi.77.2.1598-1603.2003. J Virol. 2003. PMID: 12502875 Free PMC article.
-
Nucleolar protein trafficking in response to HIV-1 Tat: rewiring the nucleolus.PLoS One. 2012;7(11):e48702. doi: 10.1371/journal.pone.0048702. Epub 2012 Nov 15. PLoS One. 2012. PMID: 23166591 Free PMC article.
-
The DNA-bending protein HMGB1 is a cellular cofactor of Sleeping Beauty transposition.Nucleic Acids Res. 2003 May 1;31(9):2313-22. doi: 10.1093/nar/gkg341. Nucleic Acids Res. 2003. PMID: 12711676 Free PMC article.
References
-
- Brown P O, Bowerman B, Varmus H E, Bishop J M. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

