Substrate binding in vitro and kinetics of RsrI [N6-adenine] DNA methyltransferase
- PMID: 11024176
- PMCID: PMC110777
- DOI: 10.1093/nar/28.20.3962
Substrate binding in vitro and kinetics of RsrI [N6-adenine] DNA methyltransferase
Abstract
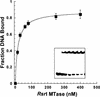
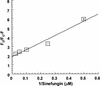
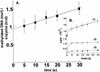
RSR:I [N:6-adenine] DNA methyltransferase (M.RSR:I), which recognizes GAATTC and is a member of a restriction-modification system in Rhodobacter sphaeroides, was purified to >95% homogeneity using a simplified procedure involving two ion exchange chromatographic steps. Electrophoretic gel retardation assays with purified M.RSR:I were performed on unmethylated, hemimethylated, dimethylated or non-specific target DNA duplexes (25 bp) in the presence of sinefungin, a potent inhibitory analog of AdoMet. M. RSR:I binding was affected by the methylation status of the DNA substrate and was enhanced by the presence of the cofactor analog. M. RSR:I bound DNA substrates in the presence of sinefungin with decreasing affinities: hemimethylated > unmethylated > dimethylated >> non-specific DNA. Gel retardation studies with DNA substrates containing an abasic site substituted for the target adenine DNA provided evidence consistent with M.RSR:I extruding the target base from the duplex. Consistent with such base flipping, an approximately 1.7-fold fluorescence intensity increase was observed upon stoichiometric addition of M.RSR:I to hemimethylated DNA containing the fluorescent analog 2-aminopurine in place of the target adenine. Pre-steady-state kinetic and isotope- partitioning experiments revealed that the enzyme displays burst kinetics, confirmed the catalytic competence of the M.RSR:I-AdoMet complex and eliminated the possibility of an ordered mechanism where DNA is required to bind first. The equilibrium dissociation constants for AdoMet, AdoHcy and sinefungin were determined using an intrinsic tryptophan fluorescence-quenching assay.
Figures

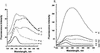
Similar articles
-
A dual role for substrate S-adenosyl-L-methionine in the methylation reaction with bacteriophage T4 Dam DNA-[N6-adenine]-methyltransferase.Nucleic Acids Res. 2001 Jun 1;29(11):2361-9. doi: 10.1093/nar/29.11.2361. Nucleic Acids Res. 2001. PMID: 11376154 Free PMC article.
-
Molecular enzymology of the EcoRV DNA-(Adenine-N (6))-methyltransferase: kinetics of DNA binding and bending, kinetic mechanism and linear diffusion of the enzyme on DNA.J Mol Biol. 2000 Oct 13;303(1):93-110. doi: 10.1006/jmbi.2000.4127. J Mol Biol. 2000. PMID: 11021972
-
Structure of RsrI methyltransferase, a member of the N6-adenine beta class of DNA methyltransferases.Nucleic Acids Res. 2000 Oct 15;28(20):3950-61. doi: 10.1093/nar/28.20.3950. Nucleic Acids Res. 2000. PMID: 11024175 Free PMC article.
-
M.TaqI: possible catalysis via cation-pi interactions in N-specific DNA methyltransferases.Biol Chem. 1998 Apr-May;379(4-5):389-400. Biol Chem. 1998. PMID: 9628329 Review.
-
Design and Application of 6mA-Specific Zinc-Finger Proteins for the Readout of DNA Methylation.Methods Mol Biol. 2018;1867:29-41. doi: 10.1007/978-1-4939-8799-3_3. Methods Mol Biol. 2018. PMID: 30155813 Review.
Cited by
-
A dual role for substrate S-adenosyl-L-methionine in the methylation reaction with bacteriophage T4 Dam DNA-[N6-adenine]-methyltransferase.Nucleic Acids Res. 2001 Jun 1;29(11):2361-9. doi: 10.1093/nar/29.11.2361. Nucleic Acids Res. 2001. PMID: 11376154 Free PMC article.
-
Nucleotide flipping by restriction enzymes analyzed by 2-aminopurine steady-state fluorescence.Nucleic Acids Res. 2007;35(14):4792-9. doi: 10.1093/nar/gkm513. Epub 2007 Jul 7. Nucleic Acids Res. 2007. PMID: 17617640 Free PMC article.
-
Structure, function and mechanism of exocyclic DNA methyltransferases.Biochem J. 2006 Oct 15;399(2):177-90. doi: 10.1042/BJ20060854. Biochem J. 2006. PMID: 16987108 Free PMC article. Review.
-
Kinetic and catalytic properties of M.HpyAXVII, a phase-variable DNA methyltransferase from Helicobacter pylori.J Biol Chem. 2019 Jan 18;294(3):1019-1034. doi: 10.1074/jbc.RA118.003769. Epub 2018 Nov 26. J Biol Chem. 2019. PMID: 30478171 Free PMC article.
-
Unusual 2-aminopurine fluorescence from a complex of DNA and the EcoKI methyltransferase.Nucleic Acids Res. 2004 Apr 23;32(7):2223-30. doi: 10.1093/nar/gkh531. Print 2004. Nucleic Acids Res. 2004. PMID: 15107490 Free PMC article.
References
-
- Cheng X. and Blumenthal,R.M. (1999) S-Adenosylmethionine-dependent Methyltransferases: Structures and Functions. World Scientific, Singapore, Singapore.
-
- Jackson-Grusby L. and Jaenisch,R. (1996) Semin. Cancer Biol., 7, 261–268. - PubMed
-
- Palmer B.R. and Marinus,M.G. (1994) Gene, 143, 1–12. - PubMed
-
- Malone T., Blumenthal,R.M. and Cheng,X. (1995) J. Mol. Biol., 253, 618–632. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

