Structure of RsrI methyltransferase, a member of the N6-adenine beta class of DNA methyltransferases
- PMID: 11024175
- PMCID: PMC110776
- DOI: 10.1093/nar/28.20.3950
Structure of RsrI methyltransferase, a member of the N6-adenine beta class of DNA methyltransferases
Abstract
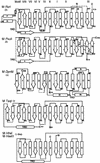
DNA methylation is important in cellular, developmental and disease processes, as well as in bacterial restriction-modification systems. Methylation of DNA at the amino groups of cytosine and adenine is a common mode of protection against restriction endonucleases afforded by the bacterial methyltransferases. The first structure of an N:6-adenine methyltransferase belonging to the beta class of bacterial methyltransferases is described here. The structure of M. RSR:I from Rhodobacter sphaeroides, which methylates the second adenine of the GAATTC sequence, was determined to 1.75 A resolution using X-ray crystallography. Like other methyltransferases, the enzyme contains the methylase fold and has well-defined substrate binding pockets. The catalytic core most closely resembles the PVU:II methyltransferase, a cytosine amino methyltransferase of the same beta group. The larger nucleotide binding pocket observed in M. RSR:I is expected because it methylates adenine. However, the most striking difference between the RSR:I methyltransferase and the other bacterial enzymes is the structure of the putative DNA target recognition domain, which is formed in part by two helices on an extended arm of the protein on the face of the enzyme opposite the active site. This observation suggests that a dramatic conformational change or oligomerization may take place during DNA binding and methylation.
Figures
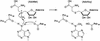

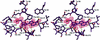


Similar articles
-
Substrate binding in vitro and kinetics of RsrI [N6-adenine] DNA methyltransferase.Nucleic Acids Res. 2000 Oct 15;28(20):3962-71. doi: 10.1093/nar/28.20.3962. Nucleic Acids Res. 2000. PMID: 11024176 Free PMC article.
-
DNA binding properties in vivo and target recognition domain sequence alignment analyses of wild-type and mutant RsrI [N6-adenine] DNA methyltransferases.Nucleic Acids Res. 2000 Oct 15;28(20):3972-81. doi: 10.1093/nar/28.20.3972. Nucleic Acids Res. 2000. PMID: 11024177 Free PMC article.
-
Structure and function of DNA methyltransferases.Annu Rev Biophys Biomol Struct. 1995;24:293-318. doi: 10.1146/annurev.bb.24.060195.001453. Annu Rev Biophys Biomol Struct. 1995. PMID: 7663118 Review.
-
Crystal structure of MboIIA methyltransferase.Nucleic Acids Res. 2003 Sep 15;31(18):5440-8. doi: 10.1093/nar/gkg713. Nucleic Acids Res. 2003. PMID: 12954781 Free PMC article.
-
M.TaqI: possible catalysis via cation-pi interactions in N-specific DNA methyltransferases.Biol Chem. 1998 Apr-May;379(4-5):389-400. Biol Chem. 1998. PMID: 9628329 Review.
Cited by
-
The phage growth limitation system in Streptomyces coelicolor A(3)2 is a toxin/antitoxin system, comprising enzymes with DNA methyltransferase, protein kinase and ATPase activity.Virology. 2015 Mar;477:100-109. doi: 10.1016/j.virol.2014.12.036. Epub 2015 Jan 13. Virology. 2015. PMID: 25592393 Free PMC article.
-
Structural insight into human N6amt1-Trm112 complex functioning as a protein methyltransferase.Cell Discov. 2019 Sep 10;5:51. doi: 10.1038/s41421-019-0121-y. eCollection 2019. Cell Discov. 2019. PMID: 31636962 Free PMC article.
-
The crystal structure of spermidine synthase with a multisubstrate adduct inhibitor.Nat Struct Biol. 2002 Jan;9(1):27-31. doi: 10.1038/nsb737. Nat Struct Biol. 2002. PMID: 11731804 Free PMC article.
-
Structural insights into DNA N6-adenine methylation by the MTA1 complex.Cell Discov. 2023 Jan 20;9(1):8. doi: 10.1038/s41421-022-00516-w. Cell Discov. 2023. PMID: 36658132 Free PMC article.
-
Beta class amino methyltransferases from bacteria to humans: evolution and structural consequences.Nucleic Acids Res. 2020 Oct 9;48(18):10034-10044. doi: 10.1093/nar/gkaa446. Nucleic Acids Res. 2020. PMID: 32453412 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

