Human nonsyndromic hereditary deafness DFNA17 is due to a mutation in nonmuscle myosin MYH9
- PMID: 11023810
- PMCID: PMC1288554
- DOI: 10.1016/S0002-9297(07)62942-5
Human nonsyndromic hereditary deafness DFNA17 is due to a mutation in nonmuscle myosin MYH9
Abstract
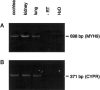
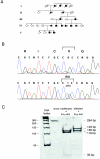
The authors had previously mapped a new locus-DFNA17, for nonsyndromic hereditary hearing impairment-to chromosome 22q12.2-q13. 3. DFNA17 spans a 17- to 23-cM region, and MYH9, a nonmuscle-myosin heavy-chain gene, is located within the linked region. Because of the importance of myosins in hearing, MYH9 was tested as a candidate gene for DFNA17. Expression of MYH9 in the rat cochlea was confirmed using reverse transcriptase-PCR and immunohistochemistry. MYH9 was immunolocalized in the organ of Corti, the subcentral region of the spiral ligament, and the Reissner membrane. Sequence analysis of MYH9 in a family with DFNA17 identified, at nucleotide 2114, a G-->A transposition that cosegregated with the inherited autosomal dominant hearing impairment. This missense mutation changes codon 705 from an invariant arginine (R) to histidine (H), R705H, within a highly conserved SH1 linker region. Previous studies have shown that modification of amino acid residues within the SH1 helix causes dysfunction of the ATPase activity of the motor domain in myosin II. Both the precise role of MYH9 in the cochlea and the mechanism by which the R705H mutation leads to the DFNA17 phenotype (progressive hearing impairment and cochleosaccular degeneration) remain to be elucidated.
Figures

Similar articles
-
Mutations in MYH9 exons 1, 16, 26, and 30 are infrequently found in Japanese patients with nonsyndromic deafness.Genet Test Mol Biomarkers. 2009 Oct;13(5):705-7. doi: 10.1089/gtmb.2009.0044. Genet Test Mol Biomarkers. 2009. PMID: 19645626
-
Absence of hearing loss in a mouse model for DFNA17 and MYH9-related disease: the use of public gene-targeted ES cell resources.Brain Res. 2006 May 26;1091(1):235-42. doi: 10.1016/j.brainres.2006.03.032. Epub 2006 Apr 21. Brain Res. 2006. PMID: 16630581
-
R705H mutation of MYH9 is associated with MYH9-related disease and not only with non-syndromic deafness DFNA17.Clin Genet. 2015 Jul;88(1):85-9. doi: 10.1111/cge.12438. Epub 2014 Jul 26. Clin Genet. 2015. PMID: 24890873
-
Advances in the understanding of MYH9 disorders.Curr Opin Hematol. 2010 Sep;17(5):405-10. doi: 10.1097/MOH.0b013e32833c069c. Curr Opin Hematol. 2010. PMID: 20601875 Review.
-
Unconventional myosins and the genetics of hearing loss.Am J Med Genet. 1999 Sep 24;89(3):147-57. doi: 10.1002/(sici)1096-8628(19990924)89:3<147::aid-ajmg5>3.0.co;2-6. Am J Med Genet. 1999. PMID: 10704189 Review.
Cited by
-
NMII forms a contractile transcellular sarcomeric network to regulate apical cell junctions and tissue geometry.Curr Biol. 2013 Apr 22;23(8):731-6. doi: 10.1016/j.cub.2013.03.039. Epub 2013 Apr 4. Curr Biol. 2013. PMID: 23562268 Free PMC article.
-
Mice deficient for the type II transmembrane serine protease, TMPRSS1/hepsin, exhibit profound hearing loss.Am J Pathol. 2007 Aug;171(2):608-16. doi: 10.2353/ajpath.2007.070068. Epub 2007 Jul 9. Am J Pathol. 2007. PMID: 17620368 Free PMC article.
-
A mutation in the gamma actin 1 (ACTG1) gene causes autosomal dominant hearing loss (DFNA20/26).J Med Genet. 2003 Dec;40(12):879-84. doi: 10.1136/jmg.40.12.879. J Med Genet. 2003. PMID: 14684684 Free PMC article.
-
Mutation spectrum and genotype-phenotype correlations in a large French cohort of MYH9-Related Disorders.Mol Genet Genomic Med. 2014 Jul;2(4):297-312. doi: 10.1002/mgg3.68. Epub 2014 Feb 7. Mol Genet Genomic Med. 2014. PMID: 25077172 Free PMC article.
-
Supporting cells eliminate dying sensory hair cells to maintain epithelial integrity in the avian inner ear.J Neurosci. 2010 Sep 15;30(37):12545-56. doi: 10.1523/JNEUROSCI.3042-10.2010. J Neurosci. 2010. PMID: 20844149 Free PMC article.
References
Electronic-Database Information
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank (for rat myh9 sequence [accession number GI 6981235])
-
- Genome Database, http://www.gdb.org
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/omim (for DFNA17 [MIM 603622])
-
- Primer3 software, http://www.genome.wi.mit.edu
References
-
- Avraham KB, Hasson T, Sobe T, Balsara B, Testa JR, Skvorak AB, Morton CC, Copeland NG, Jenkins NA (1997) Characterization of unconventional MYO6, the human homologue of the gene responsible for deafness in Snell's waltzer mice. Hum Mol Genet 6:1225–1231 - PubMed
-
- Avraham KB, Hasson T, Steel KP, Kingsley DM, Russell LB, Mooseker MS, Copeland NG, Jenkins NA (1995) The mouse Snell's waltzer deafness gene encodes an unconventional myosin required for structural integrity of inner ear hair cells. Nat Genet 11:369–375 - PubMed
-
- Collins FS (1995) Positional cloning moves from perditional to traditional. Nat Genet 9:347–350 - PubMed
-
- De Lozanne A, Spudich JA (1987) Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science 236:1086–1091 - PubMed
-
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, et al (1999) The DNA sequence of human chromosome 22. Nature 402:489–495 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

