Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p
- PMID: 11018054
- PMCID: PMC2189800
- DOI: 10.1083/jcb.151.1.69
Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p
Abstract
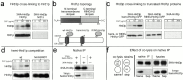
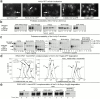
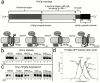
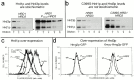
Endoplasmic reticulum (ER)-associated degradation (ERAD) is required for ubiquitin-mediated destruction of numerous proteins. ERAD occurs by processes on both sides of the ER membrane, including lumenal substrate scanning and cytosolic destruction by the proteasome. The ER resident membrane proteins Hrd1p and Hrd3p play central roles in ERAD. We show that these two proteins directly interact through the Hrd1p transmembrane domain, allowing Hrd1p stability by Hrd3p-dependent control of the Hrd1p RING-H2 domain activity. Rigorous reevaluation of Hrd1p topology demonstrated that the Hrd1p RING-H2 domain is located and functions in the cytosol. An engineered, completely lumenal, truncated version of Hrd3p functioned normally in both ERAD and Hrd1p stabilization, indicating that the lumenal domain of Hrd3p regulates the cytosolic Hrd1p RING-H2 domain by signaling through the Hrd1p transmembrane domain. Additionally, we identified a lumenal region of Hrd3p dispensable for regulation of Hrd1p stability, but absolutely required for normal ERAD. Our studies show that Hrd1p and Hrd3p form a stoichiometric complex with ERAD determinants in both the lumen and the cytosol. The HRD complex engages in lumen to cytosol communication required for regulation of Hrd1p stability and the coordination of ERAD events on both sides of the ER membrane.
Figures


Similar articles
-
Genetic interactions of Hrd3p and Der3p/Hrd1p with Sec61p suggest a retro-translocation complex mediating protein transport for ER degradation.J Cell Sci. 1999 Nov;112 ( Pt 22):4123-34. doi: 10.1242/jcs.112.22.4123. J Cell Sci. 1999. PMID: 10547371
-
Usa1p is required for optimal function and regulation of the Hrd1p endoplasmic reticulum-associated degradation ubiquitin ligase.J Biol Chem. 2010 Feb 19;285(8):5146-56. doi: 10.1074/jbc.M109.067876. Epub 2009 Nov 24. J Biol Chem. 2010. PMID: 19940128 Free PMC article.
-
Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p.Cell. 2010 Nov 12;143(4):579-91. doi: 10.1016/j.cell.2010.10.028. Cell. 2010. PMID: 21074049 Free PMC article.
-
The role of MRH domain-containing lectins in ERAD.Glycobiology. 2010 Jun;20(6):651-60. doi: 10.1093/glycob/cwq013. Epub 2010 Jan 28. Glycobiology. 2010. PMID: 20118070 Review.
-
Endoplasmic reticulum degradation: reverse protein flow of no return.FASEB J. 1997 Dec;11(14):1227-33. doi: 10.1096/fasebj.11.14.9409541. FASEB J. 1997. PMID: 9409541 Review.
Cited by
-
Selective destruction of abnormal proteins by ubiquitin-mediated protein quality control degradation.Semin Cell Dev Biol. 2012 Jul;23(5):530-7. doi: 10.1016/j.semcdb.2011.12.006. Epub 2012 Jan 8. Semin Cell Dev Biol. 2012. PMID: 22245831 Free PMC article. Review.
-
Means of self-preservation: how an intrinsically disordered ubiquitin-protein ligase averts self-destruction.Mol Biol Cell. 2013 Apr;24(7):1041-52. doi: 10.1091/mbc.E12-11-0811. Epub 2013 Jan 30. Mol Biol Cell. 2013. PMID: 23363599 Free PMC article.
-
Direct observation of autoubiquitination for an integral membrane ubiquitin ligase in ERAD.Nat Commun. 2024 Feb 13;15(1):1340. doi: 10.1038/s41467-024-45541-3. Nat Commun. 2024. PMID: 38351109 Free PMC article.
-
SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER.J Cell Biol. 2006 Oct 23;175(2):261-70. doi: 10.1083/jcb.200605196. Epub 2006 Oct 16. J Cell Biol. 2006. PMID: 17043138 Free PMC article.
-
An HRD/DER-independent ER quality control mechanism involves Rsp5p-dependent ubiquitination and ER-Golgi transport.J Cell Biol. 2002 Jul 8;158(1):91-101. doi: 10.1083/jcb.200201053. Epub 2002 Jul 8. J Cell Biol. 2002. PMID: 12105183 Free PMC article.
References
-
- Bitter G.A., Egan K.M. Expression of heterologous genes in Saccharomyces cerevisiae from vectors utilizing the glyceraldehyde-3-phosphate dehydrogenase gene promoter. Gene. 1984;32:263–274. - PubMed
-
- Bordallo J., Wolf D.H. A RING-H2 finger motif is essential for the function of Der3/Hrd1 in endoplasmic reticulum associated protein degradation in the yeast Saccharomyces cerevisiae . FEBS Lett. 1999;448:244–248. - PubMed
-
- Chun K.T., Bar-Nun S., Simoni R.D. The regulated degradation of 3-hydroxy-3-methylglutaryl-CoA reductase requires a short-lived protein and occurs in the endoplasmic reticulum. J. Biol. Chem. 1990;265:22004–22010. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

