Saccharomyces cerevisiae transcription elongation mutants are defective in PUR5 induction in response to nucleotide depletion
- PMID: 11003640
- PMCID: PMC86296
- DOI: 10.1128/MCB.20.20.7427-7437.2000
Saccharomyces cerevisiae transcription elongation mutants are defective in PUR5 induction in response to nucleotide depletion
Abstract
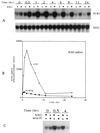
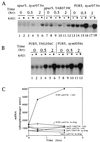
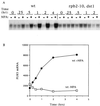
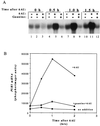
IMP dehydrogenase (IMPDH) is the rate-limiting enzyme in the de novo synthesis of guanine nucleotides. It is a target of therapeutically useful drugs and is implicated in the regulation of cell growth rate. In the yeast Saccharomyces cerevisiae, mutations in components of the RNA polymerase II (Pol II) transcription elongation machinery confer increased sensitivity to a drug that inhibits IMPDH, 6-azauracil (6AU), by a mechanism that is poorly understood. This phenotype is thought to reflect the need for an optimally functioning transcription machinery under conditions of lowered intracellular GTP levels. Here we show that in response to the application of IMPDH inhibitors such as 6AU, wild-type yeast strains induce transcription of PUR5, one of four genes encoding IMPDH-related enzymes. Yeast elongation mutants sensitive to 6AU, such as those with a disrupted gene encoding elongation factor SII or those containing amino acid substitutions in Pol II subunits, are defective in PUR5 induction. The inability to fully induce PUR5 correlates with mutations that effect transcription elongation since 6AU-sensitive strains deleted for genes not related to transcription elongation are competent to induce PUR5. DNA encompassing the PUR5 promoter and 5' untranslated region supports 6AU induction of a luciferase reporter gene in wild-type cells. Thus, yeast sense and respond to nucleotide depletion via a mechanism of transcriptional induction that restores nucleotides to levels required for normal growth. An optimally functioning elongation machinery is critical for this response.
Figures



Similar articles
-
Large-scale screening of yeast mutants for sensitivity to the IMP dehydrogenase inhibitor 6-azauracil.Yeast. 2004 Feb;21(3):241-8. doi: 10.1002/yea.1068. Yeast. 2004. PMID: 14968429 Free PMC article.
-
Regulation of an IMP dehydrogenase gene and its overexpression in drug-sensitive transcription elongation mutants of yeast.J Biol Chem. 2001 Aug 31;276(35):32905-16. doi: 10.1074/jbc.M105075200. Epub 2001 Jul 5. J Biol Chem. 2001. PMID: 11441018 Free PMC article.
-
Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II.Mol Cell Biol. 1992 Sep;12(9):4142-52. doi: 10.1128/mcb.12.9.4142-4152.1992. Mol Cell Biol. 1992. PMID: 1508210 Free PMC article.
-
Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation.Mol Cell. 2008 Jul 25;31(2):201-11. doi: 10.1016/j.molcel.2008.05.018. Mol Cell. 2008. PMID: 18657503
-
Single molecule transcription elongation.Methods. 2009 Aug;48(4):323-32. doi: 10.1016/j.ymeth.2009.04.021. Epub 2009 May 6. Methods. 2009. PMID: 19426807 Free PMC article. Review.
Cited by
-
Dissection of Pol II trigger loop function and Pol II activity-dependent control of start site selection in vivo.PLoS Genet. 2012;8(4):e1002627. doi: 10.1371/journal.pgen.1002627. Epub 2012 Apr 12. PLoS Genet. 2012. PMID: 22511879 Free PMC article.
-
Role for the Ssu72 C-terminal domain phosphatase in RNA polymerase II transcription elongation.Mol Cell Biol. 2007 Feb;27(3):926-36. doi: 10.1128/MCB.01361-06. Epub 2006 Nov 13. Mol Cell Biol. 2007. PMID: 17101794 Free PMC article.
-
TATA-binding protein mutants that are lethal in the absence of the Nhp6 high-mobility-group protein.Mol Cell Biol. 2004 Jul;24(14):6419-29. doi: 10.1128/MCB.24.14.6419-6429.2004. Mol Cell Biol. 2004. PMID: 15226442 Free PMC article.
-
Relationships Between RNA Polymerase II Activity and Spt Elongation Factors to Spt- Phenotype and Growth in Saccharomyces cerevisiae.G3 (Bethesda). 2016 Aug 9;6(8):2489-504. doi: 10.1534/g3.116.030346. G3 (Bethesda). 2016. PMID: 27261007 Free PMC article.
-
Requirement for yeast RAD26, a homolog of the human CSB gene, in elongation by RNA polymerase II.Mol Cell Biol. 2001 Dec;21(24):8651-6. doi: 10.1128/MCB.21.24.8651-8656.2001. Mol Cell Biol. 2001. PMID: 11713297 Free PMC article.
References
-
- Aparicio O M, Gottschling D E. Overcoming telomeric silencing: a trans-activator competes to establish gene expression in a cell cycle-dependent way. Genes Dev. 1994;8:1133–1146. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates/Wiley-Interscience; 1988.
-
- Barton A, Bussey H, Storms R, Kaback D. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: characterization of the 54 kb right terminal CDC14-FLO1-PHO11 region. Yeast. 1997;13:1251–1263. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
