Ric1p and Rgp1p form a complex that catalyses nucleotide exchange on Ypt6p
- PMID: 10990452
- PMCID: PMC314221
- DOI: 10.1093/emboj/19.18.4885
Ric1p and Rgp1p form a complex that catalyses nucleotide exchange on Ypt6p
Abstract
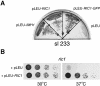
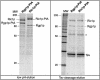
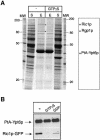
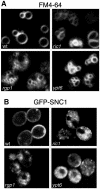
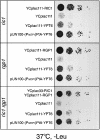
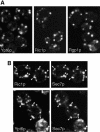
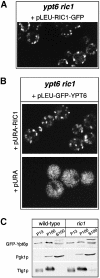
Cells lacking the GTPase Ypt6p have defects in intracellular traffic and are temperature sensitive. Their growth is severely impaired by additional mutation of IMH1, which encodes a non-essential Golgi-associated coiled-coil protein. A screen for mutants that, like ypt6, specifically impair the growth of imh1 cells led to the identification of RIC1. Ric1p forms a tight complex with a previously uncharacterized protein, Rgp1p. The Ric1p-Rgp1p complex binds Ypt6p in a nucleotide-dependent manner, and purified Ric1p-Rgp1 stimulates guanine nucleotide exchange on Ypt6p in vitro. Deletion of RIC1 or RGP1, like that of YPT6, blocks the recycling of the exocytic SNARE Snc1p from early endosomes to the Golgi and causes temperature-sensitive growth, but this defect can be relieved by overexpression of YPT6. Ric1p largely colocalizes with the late Golgi marker Sec7p. Ypt6p shows a similar distribution, but this is altered when RIC1 or RGP1 is mutated. We infer that the Ric1p-Rgp1p complex serves to activate Ypt6p on Golgi membranes by nucleotide exchange, and that this is required for efficient fusion of endosome-derived vesicles with the Golgi.
Figures

Similar articles
-
An effector of Ypt6p binds the SNARE Tlg1p and mediates selective fusion of vesicles with late Golgi membranes.EMBO J. 2001 Nov 1;20(21):5991-8. doi: 10.1093/emboj/20.21.5991. EMBO J. 2001. PMID: 11689439 Free PMC article.
-
Ric1p and the Ypt6p GTPase function in a common pathway required for localization of trans-Golgi network membrane proteins.Mol Biol Cell. 2001 Jan;12(1):13-26. doi: 10.1091/mbc.12.1.13. Mol Biol Cell. 2001. PMID: 11160819 Free PMC article.
-
The GTPase Arf1p and the ER to Golgi cargo receptor Erv14p cooperate to recruit the golgin Rud3p to the cis-Golgi.J Cell Biol. 2004 Oct 25;167(2):281-92. doi: 10.1083/jcb.200407088. J Cell Biol. 2004. PMID: 15504911 Free PMC article.
-
An acidic sequence of a putative yeast Golgi membrane protein binds COPII and facilitates ER export.EMBO J. 2001 Dec 3;20(23):6742-50. doi: 10.1093/emboj/20.23.6742. EMBO J. 2001. PMID: 11726510 Free PMC article.
-
Human Rer1 is localized to the Golgi apparatus and complements the deletion of the homologous Rer1 protein of Saccharomyces cerevisiae.Eur J Cell Biol. 1997 Sep;74(1):31-40. Eur J Cell Biol. 1997. PMID: 9309388
Cited by
-
Functional synergy between Rab5 effector Rabaptin-5 and exchange factor Rabex-5 when physically associated in a complex.Mol Biol Cell. 2001 Jul;12(7):2219-28. doi: 10.1091/mbc.12.7.2219. Mol Biol Cell. 2001. PMID: 11452015 Free PMC article.
-
Analysis of the small GTPase gene superfamily of Arabidopsis.Plant Physiol. 2003 Mar;131(3):1191-208. doi: 10.1104/pp.013052. Plant Physiol. 2003. PMID: 12644670 Free PMC article.
-
Cfs1p, a Novel Membrane Protein in the PQ-Loop Family, Is Involved in Phospholipid Flippase Functions in Yeast.G3 (Bethesda). 2017 Jan 5;7(1):179-192. doi: 10.1534/g3.116.035238. G3 (Bethesda). 2017. PMID: 28057802 Free PMC article.
-
Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p-Rcy1p pathway.Mol Biol Cell. 2007 Jan;18(1):295-312. doi: 10.1091/mbc.e06-05-0461. Epub 2006 Nov 8. Mol Biol Cell. 2007. PMID: 17093059 Free PMC article.
-
Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling.J Cell Biol. 2007 Apr 9;177(1):115-25. doi: 10.1083/jcb.200609161. J Cell Biol. 2007. PMID: 17420293 Free PMC article.
References
-
- Barr F.A. (1999) A novel Rab6-interacting domain defines a family of Golgi-targeted coiled-coil proteins. Curr. Biol., 9, 381–384. - PubMed
-
- Bensen E.S., Costaguta,G. and Payne,G.S. (2000) Synthetic genetic interactions with temperature-sensitive clathrin in Saccharomyces cerevisiae. Roles for synaptojanin-like Inp53p and dynamin-related Vps1p in clathrin-dependent protein sorting at the trans-Golgi network. Genetics, 154, 83–97. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

