Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer
- PMID: 10982342
- PMCID: PMC102094
- DOI: 10.1128/jvi.74.19.8980-8988.2000
Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer
Abstract
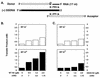
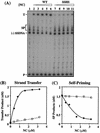
The nucleocapsid protein (NC) of human immunodeficiency virus type 1 (HIV-1) has two zinc fingers, each containing the invariant metal ion binding residues CCHC. Recent reports indicate that mutations in the CCHC motifs are deleterious for reverse transcription in vivo. To identify reverse transcriptase (RT) reactions affected by such changes, we have probed zinc finger functions in NC-dependent RT-catalyzed HIV-1 minus- and plus-strand transfer model systems. Our approach was to examine the activities of wild-type NC and a mutant in which all six cysteine residues were replaced by serine (SSHS NC); this mutation severely disrupts zinc coordination. We find that the zinc fingers contribute to the role of NC in complete tRNA primer removal from minus-strand DNA during plus-strand transfer. Annealing of the primer binding site sequences in plus-strand strong-stop DNA [(+) SSDNA] to its complement in minus-strand acceptor DNA is not dependent on NC zinc fingers. In contrast, the rate of annealing of the complementary R regions in (-) SSDNA and 3' viral RNA during minus-strand transfer is approximately eightfold lower when SSHS NC is used in place of wild-type NC. Moreover, unlike wild-type NC, SSHS NC has only a small stimulatory effect on minus-strand transfer and is essentially unable to block TAR-induced self-priming from (-) SSDNA. Our results strongly suggest that NC zinc finger structures are needed to unfold highly structured RNA and DNA strand transfer intermediates. Thus, it appears that in these cases, zinc finger interactions are important components of NC nucleic acid chaperone activity.
Figures

Similar articles
-
Subtle alterations of the native zinc finger structures have dramatic effects on the nucleic acid chaperone activity of human immunodeficiency virus type 1 nucleocapsid protein.J Virol. 2002 May;76(9):4370-8. doi: 10.1128/jvi.76.9.4370-4378.2002. J Virol. 2002. PMID: 11932404 Free PMC article.
-
Zinc finger-dependent HIV-1 nucleocapsid protein-TAR RNA interactions.Nucleic Acids Res. 2003 Aug 15;31(16):4847-55. doi: 10.1093/nar/gkg679. Nucleic Acids Res. 2003. PMID: 12907727 Free PMC article.
-
Alteration of nucleic acid structure and stability modulates the efficiency of minus-strand transfer mediated by the HIV-1 nucleocapsid protein.J Biol Chem. 2004 Oct 15;279(42):44154-65. doi: 10.1074/jbc.M401646200. Epub 2004 Jul 22. J Biol Chem. 2004. PMID: 15271979
-
Strand transfer events during HIV-1 reverse transcription.Virus Res. 2008 Jun;134(1-2):19-38. doi: 10.1016/j.virusres.2007.12.017. Epub 2008 Feb 14. Virus Res. 2008. PMID: 18279992 Review.
-
Nucleocapsid protein function in early infection processes.Virus Res. 2008 Jun;134(1-2):39-63. doi: 10.1016/j.virusres.2007.12.006. Epub 2008 Feb 14. Virus Res. 2008. PMID: 18279991 Free PMC article. Review.
Cited by
-
Site-selective probing of cTAR destabilization highlights the necessary plasticity of the HIV-1 nucleocapsid protein to chaperone the first strand transfer.Nucleic Acids Res. 2013 May;41(9):5036-48. doi: 10.1093/nar/gkt164. Epub 2013 Mar 19. Nucleic Acids Res. 2013. PMID: 23511968 Free PMC article.
-
Reverse Transcription of Retroviruses and LTR Retrotransposons.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0027-2014. doi: 10.1128/microbiolspec.MDNA3-0027-2014. Microbiol Spectr. 2015. PMID: 26104704 Free PMC article. Review.
-
Complex interactions of HIV-1 nucleocapsid protein with oligonucleotides.Nucleic Acids Res. 2006 Jan 24;34(2):472-84. doi: 10.1093/nar/gkj442. Print 2006. Nucleic Acids Res. 2006. PMID: 16434700 Free PMC article.
-
Development of a cell-based assay probing the specific interaction between the human immunodeficiency virus type 1 nucleocapsid and psi RNA in vivo.J Virol. 2007 Jun;81(11):6151-5. doi: 10.1128/JVI.00414-07. Epub 2007 Mar 14. J Virol. 2007. PMID: 17360755 Free PMC article.
-
Probing dynamics of HIV-1 nucleocapsid protein/target hexanucleotide complexes by 2-aminopurine.Nucleic Acids Res. 2008 Feb;36(3):885-96. doi: 10.1093/nar/gkm1109. Epub 2007 Dec 17. Nucleic Acids Res. 2008. PMID: 18086707 Free PMC article.
References
-
- Arad U. Modified Hirt procedure for rapid purification of extrachromosomal DNA from mammalian cells. BioTechniques. 1998;24:760–762. - PubMed
-
- Arts E J, Wainberg M A. Human immunodeficiency virus type 1 reverse transcriptase and early events in reverse transcription. Adv Virus Res. 1996;46:97–163. - PubMed
-
- Auxilien S, Keith G, Le Grice S F J, Darlix J-L. Role of post-transcriptional modifications of primer tRNALys,3 in the fidelity and efficacy of plus-strand DNA transfer during HIV-1 reverse transcription. J Biol Chem. 1999;274:4412–4420. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

