Bacterial and yeast chaperones reduce both aggregate formation and cell death in mammalian cell models of Huntington's disease
- PMID: 10920207
- PMCID: PMC16928
- DOI: 10.1073/pnas.170280697
Bacterial and yeast chaperones reduce both aggregate formation and cell death in mammalian cell models of Huntington's disease
Abstract
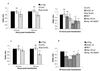
Huntington's disease (HD) is an autosomal dominant neurodegenerative condition caused by expansions of more than 35 uninterrupted CAG repeats in exon 1 of the huntingtin gene. The CAG repeats in HD and the other seven known diseases caused by CAG codon expansions are translated into long polyglutamine tracts that confer a deleterious gain of function on the mutant proteins. Intraneuronal inclusions comprising aggregates of the relevant mutant proteins are found in the brains of patients with HD and related diseases. It is crucial to determine whether the formation of inclusions is directly pathogenic, because a number of studies have suggested that aggregates may be epiphenomena or even protective. Here, we show that fragments of the bacterial chaperone GroEL and the full-length yeast heat shock protein Hsp104 reduce both aggregate formation and cell death in mammalian cell models of HD, consistent with a causal link between aggregation and pathology.
Figures
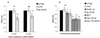
Similar articles
-
The bacterial chaperonin GroEL requires GroES to reduce aggregation and cell death in a COS-7 cell model of Huntington's disease.Neurosci Lett. 2002 Sep 27;330(3):270-4. doi: 10.1016/s0304-3940(02)00770-x. Neurosci Lett. 2002. PMID: 12270644
-
Time-lapse analysis of aggregate formation in an inducible PC12 cell model of Huntington's disease reveals time-dependent aggregate formation that transiently delays cell death.Brain Res Bull. 2008 Jan 31;75(1):146-57. doi: 10.1016/j.brainresbull.2007.08.005. Epub 2007 Sep 17. Brain Res Bull. 2008. PMID: 18158109
-
Structure and function of the molecular chaperone Hsp104 from yeast.Biopolymers. 2010 Mar;93(3):252-76. doi: 10.1002/bip.21301. Biopolymers. 2010. PMID: 19768774 Review.
-
Global changes to the ubiquitin system in Huntington's disease.Nature. 2007 Aug 9;448(7154):704-8. doi: 10.1038/nature06022. Nature. 2007. PMID: 17687326
-
Selective neuronal degeneration in Huntington's disease.Curr Top Dev Biol. 2006;75:25-71. doi: 10.1016/S0070-2153(06)75002-5. Curr Top Dev Biol. 2006. PMID: 16984809 Review.
Cited by
-
Two novel DnaJ chaperone proteins CG5001 and P58IPK regulate the pathogenicity of Huntington's disease related aggregates.Sci Rep. 2024 Sep 6;14(1):20867. doi: 10.1038/s41598-024-71065-3. Sci Rep. 2024. PMID: 39242711 Free PMC article.
-
Physical chemistry of polyglutamine: intriguing tales of a monotonous sequence.J Mol Biol. 2012 Aug 24;421(4-5):466-90. doi: 10.1016/j.jmb.2012.01.030. Epub 2012 Jan 27. J Mol Biol. 2012. PMID: 22306404 Free PMC article. Review.
-
Aggregation kinetics of interrupted polyglutamine peptides.J Mol Biol. 2011 Sep 23;412(3):505-19. doi: 10.1016/j.jmb.2011.07.003. Epub 2011 Jul 29. J Mol Biol. 2011. PMID: 21821045 Free PMC article.
-
Huntington's Disease.Cold Spring Harb Perspect Biol. 2011 Jun 1;3(6):a007476. doi: 10.1101/cshperspect.a007476. Cold Spring Harb Perspect Biol. 2011. PMID: 21441583 Free PMC article. Review.
-
Protein aggregation and polyasparagine-mediated cellular toxicity in Saccharomyces cerevisiae.Prion. 2007 Apr-Jun;1(2):144-53. doi: 10.4161/pri.1.2.4630. Epub 2007 Apr 26. Prion. 2007. PMID: 19164913 Free PMC article.
References
-
- The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. - PubMed
-
- Davies S W, Turmaine M, Cozens B A, DiFiglia M, Sharp A H, Ross C A, Scherzinger E, Wanker E E, Mangiarini L, Bates G P. Cell. 1997;90:537–548. - PubMed
-
- DiFiglia M, Sapp E, Chase K O, Davies S W, Bates G P, Vonsattel J P, Aronin N. Science. 1997;277:1990–1993. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

