NMR structures of three single-residue variants of the human prion protein
- PMID: 10900000
- PMCID: PMC26949
- DOI: 10.1073/pnas.97.15.8340
NMR structures of three single-residue variants of the human prion protein
Abstract
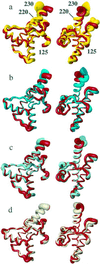
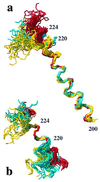
The NMR structures of three single-amino acid variants of the C-terminal domain of the human prion protein, hPrP(121-230), are presented. In hPrP(M166V) and hPrP(R220K) the substitution is with the corresponding residue in murine PrP, and in hPrP(S170N) it is with the corresponding Syrian hamster residue. All three substitutions are in the surface region of the structure of the cellular form of PrP (PrP(C)) that is formed by the C-terminal part of helix 3, with residues 218-230, and a loop of residues 166-172. This molecular region shows high species variability and has been implicated in specific interactions with a so far not further characterized "protein X," and it is related to the species barrier for transmission of prion diseases. As expected, the three variant hPrP(121-230) structures have the same global architecture as the previously determined wild-type bovine, human, murine, and Syrian hamster prion proteins, but with the present study two localized "conformational markers" could be related with single amino acid exchanges. These are the length and quality of definition of helix 3, and the NMR-observability of the residues in the loop 166-172. Poor definition of the C-terminal part of helix 3 is characteristic for murine PrP and has now been observed also for hPrP(R220K), and NMR observation of the complete loop 166-172 has so far been unique for Syrian hamster PrP and is now also documented for hPrP(S170N).
Figures


Similar articles
-
NMR solution structure of the human prion protein.Proc Natl Acad Sci U S A. 2000 Jan 4;97(1):145-50. doi: 10.1073/pnas.97.1.145. Proc Natl Acad Sci U S A. 2000. PMID: 10618385 Free PMC article.
-
NMR structure of the bank vole prion protein at 20 degrees C contains a structured loop of residues 165-171.J Mol Biol. 2008 Nov 7;383(2):306-12. doi: 10.1016/j.jmb.2008.08.045. Epub 2008 Aug 26. J Mol Biol. 2008. PMID: 18773909
-
NMR structure of the bovine prion protein.Proc Natl Acad Sci U S A. 2000 Jul 18;97(15):8334-9. doi: 10.1073/pnas.97.15.8334. Proc Natl Acad Sci U S A. 2000. PMID: 10899999 Free PMC article.
-
Molecular biology of prions causing infectious and genetic encephalopathies of humans as well as scrapie of sheep and BSE of cattle.Dev Biol Stand. 1991;75:55-74. Dev Biol Stand. 1991. PMID: 1686599 Review.
-
Prion encephalopathies of animals and humans.Dev Biol Stand. 1993;80:31-44. Dev Biol Stand. 1993. PMID: 8270114 Review.
Cited by
-
Evidence for assembly of prions with left-handed beta-helices into trimers.Proc Natl Acad Sci U S A. 2004 Jun 1;101(22):8342-7. doi: 10.1073/pnas.0402254101. Epub 2004 May 21. Proc Natl Acad Sci U S A. 2004. PMID: 15155909 Free PMC article.
-
Prion protein NMR structures of cats, dogs, pigs, and sheep.Proc Natl Acad Sci U S A. 2005 Jan 18;102(3):640-5. doi: 10.1073/pnas.0408937102. Epub 2005 Jan 12. Proc Natl Acad Sci U S A. 2005. PMID: 15647367 Free PMC article.
-
Unique structural characteristics of the rabbit prion protein.J Biol Chem. 2010 Oct 8;285(41):31682-93. doi: 10.1074/jbc.M110.118844. Epub 2010 Jul 16. J Biol Chem. 2010. PMID: 20639199 Free PMC article.
-
Prion Diseases: Update on Mad Cow Disease, Variant Creutzfeldt-Jakob Disease, and the Transmissible Spongiform Encephalopathies.Curr Infect Dis Rep. 2004 Aug;6(4):305-315. doi: 10.1007/s11908-004-0053-y. Curr Infect Dis Rep. 2004. PMID: 15265460
-
Predicted consequences of site-directed mutagenesis and the impact of species variation on prion protein misfolding through the N-terminal domain.J Mol Model. 2005 Nov;11(6):468-73. doi: 10.1007/s00894-005-0239-8. Epub 2005 Jul 21. J Mol Model. 2005. PMID: 16034619
References
-
- Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wüthrich K. Nature (London) 1996;382:180–182. - PubMed
-
- Riek R, Hornemann S, Wider G, Glockshuber R, Wüthrich K. FEBS Lett. 1997;413:282–288. - PubMed
-
- Liu H, Farr-Jones S, Ulyanov N B, Llinas M, Marqusee S, Groth D, Cohen F E, Prusiner S B, James T L. Biochemistry. 1999;38:5362–5377. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

