HMG-I/Y, a new c-Myc target gene and potential oncogene
- PMID: 10891489
- PMCID: PMC86000
- DOI: 10.1128/MCB.20.15.5490-5502.2000
HMG-I/Y, a new c-Myc target gene and potential oncogene
Abstract
The HMG-I/Y gene encodes the HMG-I and HMG-Y proteins, which function as architectural chromatin binding proteins important in the transcriptional regulation of several genes. Although increased expression of the HMG-I/Y proteins is associated with cellular proliferation, neoplastic transformation, and several human cancers, the role of these proteins in the pathogenesis of malignancy remains unclear. To better understand the role of these proteins in cell growth and transformation, we have been studying the regulation and function of HMG-I/Y. The HMG-I/Y promoter was cloned, sequenced, and subjected to mutagenesis analysis. A c-Myc-Max consensus DNA binding site was identified as an element important in the serum stimulation of HMG-I/Y. The oncoprotein c-Myc and its protein partner Max bind to this site in vitro and activate transcription in transfection experiments. HMG-I/Y expression is stimulated by c-Myc in a Myc-estradiol receptor cell line in the presence of the protein synthesis inhibitor cycloheximide, indicating that HMG-I/Y is a direct c-Myc target gene. HMG-I/Y induction is decreased in Myc-deficient fibroblasts. HMG-I/Y protein expression is also increased in Burkitt's lymphoma cell lines, which are known to have increased c-Myc protein. Like Myc, increased expression of HMG-I protein leads to the neoplastic transformation of both Rat 1a fibroblasts and CB33 cells. In addition, Rat 1a cells overexpressing HMG-I protein form tumors in nude mice. Decreasing HMG-I/Y proteins using an antisense construct abrogates transformation in Burkitt's lymphoma cells. These findings indicate that HMG-I/Y is a c-Myc target gene involved in neoplastic transformation and a member of a new class of potential oncogenes.
Figures
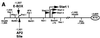
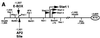
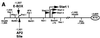
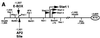






Similar articles
-
The oncogenic properties of the HMG-I gene family.Cancer Res. 2000 Aug 1;60(15):4256-61. Cancer Res. 2000. PMID: 10945639
-
A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells.Proc Natl Acad Sci U S A. 2003 Jul 8;100(14):8164-9. doi: 10.1073/pnas.1332764100. Epub 2003 Jun 13. Proc Natl Acad Sci U S A. 2003. PMID: 12808131 Free PMC article.
-
The tmp gene, encoding a membrane protein, is a c-Myc target with a tumorigenic activity.Mol Cell Biol. 1999 May;19(5):3529-39. doi: 10.1128/MCB.19.5.3529. Mol Cell Biol. 1999. PMID: 10207076 Free PMC article.
-
Target genes and cellular regulators of the Myc transcription complex.Crit Rev Eukaryot Gene Expr. 1998;8(3-4):277-96. doi: 10.1615/critreveukargeneexpr.v8.i3-4.30. Crit Rev Eukaryot Gene Expr. 1998. PMID: 9807697 Review.
-
The role of c-myc in cell growth.Curr Opin Genet Dev. 1993 Feb;3(1):44-9. doi: 10.1016/s0959-437x(05)80339-9. Curr Opin Genet Dev. 1993. PMID: 8453273 Review.
Cited by
-
HMGA1: a master regulator of tumor progression in triple-negative breast cancer cells.PLoS One. 2013 May 2;8(5):e63419. doi: 10.1371/journal.pone.0063419. Print 2013. PLoS One. 2013. PMID: 23658826 Free PMC article.
-
Lentivirus-mediated RNA interference of HMGA1 promotes chemosensitivity to gemcitabine in pancreatic adenocarcinoma.J Gastrointest Surg. 2006 Nov;10(9):1254-62; discussion 1263. doi: 10.1016/j.gassur.2006.06.011. J Gastrointest Surg. 2006. PMID: 17114012
-
Inhibition of high-mobility-group A2 protein binding to DNA by netropsin: a biosensor-surface plasmon resonance assay.Anal Biochem. 2008 Mar 1;374(1):7-15. doi: 10.1016/j.ab.2007.10.023. Epub 2007 Oct 23. Anal Biochem. 2008. PMID: 18023407 Free PMC article.
-
BCL2 translocation defines a unique tumor subset within the germinal center B-cell-like diffuse large B-cell lymphoma.Am J Pathol. 2004 Jul;165(1):159-66. doi: 10.1016/s0002-9440(10)63284-1. Am J Pathol. 2004. PMID: 15215171 Free PMC article.
-
Anti-Transcription Factor RNA Aptamers as Potential Therapeutics.Nucleic Acid Ther. 2016 Feb;26(1):29-43. doi: 10.1089/nat.2015.0566. Epub 2015 Oct 28. Nucleic Acid Ther. 2016. PMID: 26509637 Free PMC article. Review.
References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1990.
-
- Auvinen M, Laine A, Paasinen-Sohns A, Kangas A, Kangas L, Saksela O, Andersson L C, Holtta E. Human ornithine decarboxylase-overproducing NIH3T3 cells induce rapidly growing, highly vascularized tumors in nude mice. Cancer Res. 1997;57:3016–3025. - PubMed
-
- Auvinen M, Paasinen A, Andersson L C, Holtta E. Ornithine decarboxylase activity is critical for cell transformation. Nature. 1992;360:355–358. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
