Characterization of an essential RNA secondary structure in the 3' untranslated region of the murine coronavirus genome
- PMID: 10888630
- PMCID: PMC112208
- DOI: 10.1128/jvi.74.15.6911-6921.2000
Characterization of an essential RNA secondary structure in the 3' untranslated region of the murine coronavirus genome
Abstract
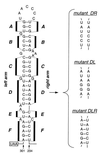
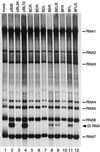
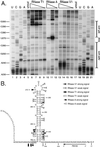
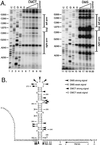
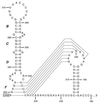
We have previously identified a functionally essential bulged stem-loop in the 3' untranslated region of the positive-stranded RNA genome of mouse hepatitis virus. This 68-nucleotide structure is composed of six stem segments interrupted by five bulges, and its structure, but not its primary sequence, is entirely conserved in the related bovine coronavirus. The functional importance of individual stem segments of this stem-loop was characterized by genetic analysis using targeted RNA recombination. We also examined the effects of stem segment mutations on the replication of mouse hepatitis virus defective interfering RNAs. These studies were complemented by enzymatic and chemical probing of the stem-loop. Taken together, our results confirmed most of the previously proposed structure, but they revealed that the terminal loop and an internal loop are larger than originally thought. Three of the stem segments were found to be essential for viral replication. Further, our results suggest that the stem segment at the base of the stem-loop is an alternative base-pairing structure for part of a downstream, and partially overlapping, RNA pseudoknot that has recently been shown to be necessary for bovine coronavirus replication.
Figures


Similar articles
-
Characterization of the RNA components of a putative molecular switch in the 3' untranslated region of the murine coronavirus genome.J Virol. 2004 Jan;78(2):669-82. doi: 10.1128/jvi.78.2.669-682.2004. J Virol. 2004. PMID: 14694098 Free PMC article.
-
Genetic interactions between an essential 3' cis-acting RNA pseudoknot, replicase gene products, and the extreme 3' end of the mouse coronavirus genome.J Virol. 2008 Feb;82(3):1214-28. doi: 10.1128/JVI.01690-07. Epub 2007 Nov 21. J Virol. 2008. PMID: 18032506 Free PMC article.
-
Secondary structural elements within the 3' untranslated region of mouse hepatitis virus strain JHM genomic RNA.J Virol. 2001 Dec;75(24):12105-13. doi: 10.1128/JVI.75.24.12105-12113.2001. J Virol. 2001. PMID: 11711601 Free PMC article.
-
[Structure and function of the non-coding regions of hepatitis C viral RNA].Postepy Biochem. 2006;52(1):62-71. Postepy Biochem. 2006. PMID: 16869303 Review. Polish.
-
The amazing diversity of cap-independent translation elements in the 3'-untranslated regions of plant viral RNAs.Biochem Soc Trans. 2007 Dec;35(Pt 6):1629-33. doi: 10.1042/BST0351629. Biochem Soc Trans. 2007. PMID: 18031280 Free PMC article. Review.
Cited by
-
Heterologous gene expression from transmissible gastroenteritis virus replicon particles.J Virol. 2002 Feb;76(3):1422-34. doi: 10.1128/jvi.76.3.1422-1434.2002. J Virol. 2002. PMID: 11773416 Free PMC article.
-
Common RNA replication signals exist among group 2 coronaviruses: evidence for in vivo recombination between animal and human coronavirus molecules.Virology. 2003 Oct 10;315(1):174-83. doi: 10.1016/s0042-6822(03)00511-7. Virology. 2003. PMID: 14592769 Free PMC article.
-
Genomic characterization of equine coronavirus.Virology. 2007 Dec 5;369(1):92-104. doi: 10.1016/j.virol.2007.06.035. Epub 2007 Aug 13. Virology. 2007. PMID: 17706262 Free PMC article.
-
Bovine coronavirus nonstructural protein 1 (p28) is an RNA binding protein that binds terminal genomic cis-replication elements.J Virol. 2009 Jun;83(12):6087-97. doi: 10.1128/JVI.00160-09. Epub 2009 Apr 8. J Virol. 2009. PMID: 19357173 Free PMC article.
-
The coronavirus recombination pathway.Cell Host Microbe. 2023 Jun 14;31(6):874-889. doi: 10.1016/j.chom.2023.05.003. Cell Host Microbe. 2023. PMID: 37321171 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

