A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions
- PMID: 10852947
- PMCID: PMC16554
- DOI: 10.1073/pnas.110149297
A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions
Abstract
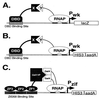
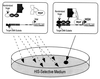
We have developed a bacterial "two-hybrid" system that readily allows selection from libraries larger than 10(8) in size. Our bacterial system may be used to study either protein-DNA or protein-protein interactions, and it offers a number of potentially significant advantages over existing yeast-based one-hybrid and two-hybrid methods. We tested our system by selecting zinc finger variants (from a large randomized library) that bind tightly and specifically to desired DNA target sites. Our method allows sequence-specific zinc fingers to be isolated in a single selection step, and thus it should be more rapid than phage display strategies that typically require multiple enrichment/amplification cycles. Given the large library sizes our bacterial-based selection system can handle, this method should provide a powerful tool for identifying and optimizing protein-DNA and protein-protein interactions.
Figures

Similar articles
-
Identifying and modifying protein-DNA and protein-protein interactions using a bacterial two-hybrid selection system.J Cell Biochem Suppl. 2001;Suppl 37:53-7. doi: 10.1002/jcb.10065. J Cell Biochem Suppl. 2001. PMID: 11842428 Review.
-
Engineering Cys2His2 zinc finger domains using a bacterial cell-based two-hybrid selection system.Methods Mol Biol. 2007;408:317-34. doi: 10.1007/978-1-59745-547-3_17. Methods Mol Biol. 2007. PMID: 18314590
-
Combining structure-based design with phage display to create new Cys(2)His(2) zinc finger dimers.Structure. 2000 Jul 15;8(7):739-50. doi: 10.1016/s0969-2126(00)00161-1. Structure. 2000. PMID: 10903945
-
Engineering single Cys2His2 zinc finger domains using a bacterial cell-based two-hybrid selection system.Methods Mol Biol. 2010;649:31-50. doi: 10.1007/978-1-60761-753-2_2. Methods Mol Biol. 2010. PMID: 20680826
-
One from column A and two from column B: the benefits of phage display in molecular-recognition studies.Curr Opin Chem Biol. 2002 Feb;6(1):92-6. doi: 10.1016/s1367-5931(01)00287-3. Curr Opin Chem Biol. 2002. PMID: 11827830 Review.
Cited by
-
Nonoptimal Codon Usage Is Critical for Protein Structure and Function of the Master General Amino Acid Control Regulator CPC-1.mBio. 2020 Oct 13;11(5):e02605-20. doi: 10.1128/mBio.02605-20. mBio. 2020. PMID: 33051373 Free PMC article.
-
Insights into the role of the junctional region of Plasmodium falciparum dihydrofolate reductase-thymidylate synthase.Malar J. 2013 Mar 12;12:91. doi: 10.1186/1475-2875-12-91. Malar J. 2013. PMID: 23497065 Free PMC article.
-
Magnitude of the CREB-dependent transcriptional response is determined by the strength of the interaction between the kinase-inducible domain of CREB and the KIX domain of CREB-binding protein.Mol Cell Biol. 2000 Dec;20(24):9409-22. doi: 10.1128/MCB.20.24.9409-9422.2000. Mol Cell Biol. 2000. PMID: 11094091 Free PMC article.
-
Highly specific zinc finger proteins obtained by directed domain shuffling and cell-based selection.Proc Natl Acad Sci U S A. 2003 Oct 14;100(21):12271-6. doi: 10.1073/pnas.2135381100. Epub 2003 Oct 3. Proc Natl Acad Sci U S A. 2003. PMID: 14527993 Free PMC article.
-
Identification of a DNA-binding site and transcriptional target for the EWS-WT1(+KTS) oncoprotein.Genes Dev. 2003 Sep 1;17(17):2094-107. doi: 10.1101/gad.1110703. Epub 2003 Aug 15. Genes Dev. 2003. PMID: 12923058 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

