Eap1p, a novel eukaryotic translation initiation factor 4E-associated protein in Saccharomyces cerevisiae
- PMID: 10848587
- PMCID: PMC85860
- DOI: 10.1128/MCB.20.13.4604-4613.2000
Eap1p, a novel eukaryotic translation initiation factor 4E-associated protein in Saccharomyces cerevisiae
Abstract
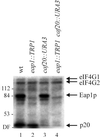
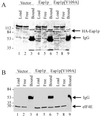
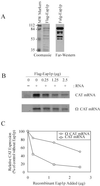
Ribosome binding to eukaryotic mRNA is a multistep process which is mediated by the cap structure [m(7)G(5')ppp(5')N, where N is any nucleotide] present at the 5' termini of all cellular (with the exception of organellar) mRNAs. The heterotrimeric complex, eukaryotic initiation factor 4F (eIF4F), interacts directly with the cap structure via the eIF4E subunit and functions to assemble a ribosomal initiation complex on the mRNA. In mammalian cells, eIF4E activity is regulated in part by three related translational repressors (4E-BPs), which bind to eIF4E directly and preclude the assembly of eIF4F. No structural counterpart to 4E-BPs exists in the budding yeast, Saccharomyces cerevisiae. However, a functional homolog (named p20) has been described which blocks cap-dependent translation by a mechanism analogous to that of 4E-BPs. We report here on the characterization of a novel yeast eIF4E-associated protein (Eap1p) which can also regulate translation through binding to eIF4E. Eap1p shares limited homology to p20 in a region which contains the canonical eIF4E-binding motif. Deletion of this domain or point mutation abolishes the interaction of Eap1p with eIF4E. Eap1p competes with eIF4G (the large subunit of the cap-binding complex, eIF4F) and p20 for binding to eIF4E in vivo and inhibits cap-dependent translation in vitro. Targeted disruption of the EAP1 gene results in a temperature-sensitive phenotype and also confers partial resistance to growth inhibition by rapamycin. These data indicate that Eap1p plays a role in cell growth and implicates this protein in the TOR signaling cascade of S. cerevisiae.
Figures






Similar articles
-
A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E.EMBO J. 1997 Mar 3;16(5):1114-21. doi: 10.1093/emboj/16.5.1114. EMBO J. 1997. PMID: 9118949 Free PMC article.
-
Repressor binding to a dorsal regulatory site traps human eIF4E in a high cap-affinity state.EMBO J. 1999 Jul 15;18(14):4068-75. doi: 10.1093/emboj/18.14.4068. EMBO J. 1999. PMID: 10406811 Free PMC article.
-
Structural motifs in eIF4G and 4E-BPs modulate their binding to eIF4E to regulate translation initiation in yeast.Nucleic Acids Res. 2018 Jul 27;46(13):6893-6908. doi: 10.1093/nar/gky542. Nucleic Acids Res. 2018. PMID: 30053226 Free PMC article.
-
eIF4E activity is regulated at multiple levels.Int J Biochem Cell Biol. 1999 Jan;31(1):43-57. doi: 10.1016/s1357-2725(98)00131-9. Int J Biochem Cell Biol. 1999. PMID: 10216943 Review.
-
eIF4E and Interactors from Unicellular Eukaryotes.Int J Mol Sci. 2020 Mar 21;21(6):2170. doi: 10.3390/ijms21062170. Int J Mol Sci. 2020. PMID: 32245232 Free PMC article. Review.
Cited by
-
Regulation of CLB6 expression by the cytoplasmic deadenylase Ccr4 through its coding and 3' UTR regions.PLoS One. 2022 May 6;17(5):e0268283. doi: 10.1371/journal.pone.0268283. eCollection 2022. PLoS One. 2022. PMID: 35522675 Free PMC article.
-
Control of transcription by cell size.PLoS Biol. 2010 Nov 2;8(11):e1000523. doi: 10.1371/journal.pbio.1000523. PLoS Biol. 2010. PMID: 21072241 Free PMC article.
-
Ksp1-dependent phosphorylation of eIF4G modulates post-transcriptional regulation of specific mRNAs under glucose deprivation conditions.Nucleic Acids Res. 2018 Apr 6;46(6):3047-3060. doi: 10.1093/nar/gky097. Nucleic Acids Res. 2018. PMID: 29438499 Free PMC article.
-
Target of rapamycin (TOR) in nutrient signaling and growth control.Genetics. 2011 Dec;189(4):1177-201. doi: 10.1534/genetics.111.133363. Genetics. 2011. PMID: 22174183 Free PMC article. Review.
-
Telomere length kinetics assay (TELKA) sorts the telomere length maintenance (tlm) mutants into functional groups.Nucleic Acids Res. 2014 Jun;42(10):6314-25. doi: 10.1093/nar/gku267. Epub 2014 Apr 11. Nucleic Acids Res. 2014. PMID: 24728996 Free PMC article.
References
-
- Altmann M, Blum S, Wilson T M, Trachsel H. The 5′-leader sequence of tobacco mosaic virus RNA mediates initiation-factor-4E-independent, but still initiation-factor-4A-dependent translation in yeast extracts. Gene. 1990;91:127–129. - PubMed
-
- Altmann M, Edery I, Sonenberg N, Trachsel H. Purification and characterization of protein synthesis initiation factor eIF-4E from the yeast Saccharomyces cerevisiae. Biochemistry. 1985;24:6085–6089. - PubMed
-
- Altmann M, Muller P P, Pelletier J, Sonenberg N, Trachsel H. A mammalian translation initiation factor can substitute for its yeast homologue in vivo. J Biol Chem. 1989;264:12145–12147. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
