The chromo domain protein chd1p from budding yeast is an ATP-dependent chromatin-modifying factor
- PMID: 10811623
- PMCID: PMC384354
- DOI: 10.1093/emboj/19.10.2323
The chromo domain protein chd1p from budding yeast is an ATP-dependent chromatin-modifying factor
Abstract
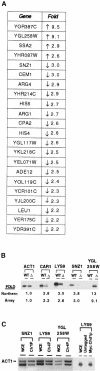
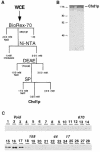
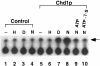
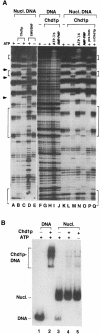
CHD proteins are members of the chromo domain family, a class of proteins involved in transcription, DNA degradation and chromatin structure. In higher eukaryotes, there are two distinct subfamilies of CHD proteins: CHD1 and CHD3/4. Analyses carried out in vitro indicate that the CHD3/4 proteins may regulate transcription via alteration of chromatin structure. However, little is known about the role of CHD proteins in vivo, particularly the CHD1 subfamily. To understand better the cellular function of CHD proteins, we initiated a study on the Chd1p protein from budding yeast. Using genomic DNA arrays, we identified genes whose expression is affected by the absence of Chd1p. A synthetic-lethal screen uncovered genetic interactions between SWI/SNF genes and CHD1. Biochemical experiments using Chd1p purified from yeast showed that it reconfigures the structure of nucleosome core particles in a manner distinct from the SWI-SNF complex. Taken together, these results suggest that Chd1p functions as a nucleosome remodeling factor, and that Chd1p may share overlapping roles with the SWI-SNF complex to regulate transcription.
Figures

Similar articles
-
Gal4-VP16 directs ATP-independent chromatin reorganization in a yeast chromatin assembly system.Biochemistry. 2005 Mar 22;44(11):4551-61. doi: 10.1021/bi047523u. Biochemistry. 2005. PMID: 15766286
-
Recruitment of chromatin remodelling factors during gene activation via the glucocorticoid receptor N-terminal domain.Biochem Soc Trans. 2000;28(4):410-4. Biochem Soc Trans. 2000. PMID: 10961930 Review.
-
Nucleosome mobilization catalysed by the yeast SWI/SNF complex.Nature. 1999 Aug 19;400(6746):784-7. doi: 10.1038/23506. Nature. 1999. PMID: 10466730
-
A study of biochemical and functional interactions of Htl1p, a putative component of the Saccharomyces cerevisiae, Rsc chromatin-remodeling complex.Gene. 2007 Jun 15;395(1-2):72-85. doi: 10.1016/j.gene.2007.02.002. Epub 2007 Feb 20. Gene. 2007. PMID: 17400406
-
The SWI-SNF complex: a chromatin remodeling machine?Trends Biochem Sci. 1995 Apr;20(4):143-6. doi: 10.1016/s0968-0004(00)88990-2. Trends Biochem Sci. 1995. PMID: 7770913 Review.
Cited by
-
The Chd family of chromatin remodelers.Mutat Res. 2007 May 1;618(1-2):30-40. doi: 10.1016/j.mrfmmm.2006.07.012. Epub 2007 Jan 21. Mutat Res. 2007. PMID: 17350655 Free PMC article. Review.
-
Retrotransposon target site selection by imitation of a cellular protein.Mol Cell Biol. 2008 Feb;28(4):1230-9. doi: 10.1128/MCB.01502-07. Epub 2007 Dec 17. Mol Cell Biol. 2008. PMID: 18086891 Free PMC article.
-
ATP-dependent chromatin remodeling: genetics, genomics and mechanisms.Cell Res. 2011 Mar;21(3):396-420. doi: 10.1038/cr.2011.32. Epub 2011 Mar 1. Cell Res. 2011. PMID: 21358755 Free PMC article. Review.
-
In Saccharomyces cerevisiae, yKu and subtelomeric core X sequences repress homologous recombination near telomeres as part of the same pathway.Genetics. 2009 Oct;183(2):441-51, 1SI-12SI. doi: 10.1534/genetics.109.106674. Epub 2009 Aug 3. Genetics. 2009. PMID: 19652177 Free PMC article.
-
CHD1 and CHD2 are positive regulators of HIV-1 gene expression.Virol J. 2014 Oct 8;11:180. doi: 10.1186/1743-422X-11-180. Virol J. 2014. PMID: 25297984 Free PMC article.
References
-
- Aparicio O.M., Weinstein,D.M. and Bell,S.P. (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell, 91, 59–69. - PubMed
-
- Cairns B.R. et al. (1996) RSC, an essential, abundant chromatin-remodeling complex. Cell, 87, 1249–1260. - PubMed
-
- Cavalli G. and Paro,R. (1998) Chromo-domain proteins: linking chromatin structure to epigenetic regulation. Curr. Opin. Cell Biol., 10, 354–360. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

