A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis
- PMID: 10781094
- PMCID: PMC18325
- DOI: 10.1073/pnas.080502497
A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis
Abstract
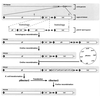
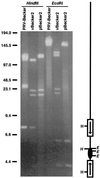
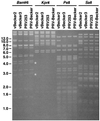
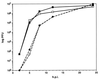
A self-recombining bacterial artificial chromosome (BAC) containing the 142-kb pseudorabies virus genome was constructed such that the viral genome is rapidly excised from the BAC vector backbone on delivery into mammalian cells. The recombination is mediated by loxP sites in the plasmid and Cre recombinase encoded within the BAC vector. A synthetic intron inserted in the middle of the cre ORF completely inhibits recombination in Escherichia coli, but is spliced out after delivery of the plasmid into mammalian cells. Recombination is efficient, and pure virus lacking the BAC vector backbone is immediately isolated from transfected mammalian cells without the need of serial passage or plaque purification.
Figures



Similar articles
-
Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene.J Virol. 2002 Mar;76(5):2316-28. doi: 10.1128/jvi.76.5.2316-2328.2002. J Virol. 2002. PMID: 11836410 Free PMC article.
-
Construction of an infectious bacterial artificial chromosome clone of a pseudorabies virus variant: Reconstituted virus exhibited wild-type properties in vitro and in vivo.J Virol Methods. 2018 Sep;259:106-115. doi: 10.1016/j.jviromet.2018.06.004. Epub 2018 Jun 9. J Virol Methods. 2018. PMID: 29894711
-
[Construction of an infectious clone of pseudorabies virus strain ZJ genome maintained as a bacterial artificial chromosome].Bing Du Xue Bao. 2010 Jul;26(4):330-5. Bing Du Xue Bao. 2010. PMID: 20836388 Chinese.
-
New tools to convert bacterial artificial chromosomes to a self-excising design and their application to a herpes simplex virus type 1 infectious clone.BMC Biotechnol. 2016 Aug 31;16(1):64. doi: 10.1186/s12896-016-0295-4. BMC Biotechnol. 2016. PMID: 27580861 Free PMC article.
-
Genetic engineering of herpes simplex virus and vector genomes carrying loxP sites in cells expressing Cre recombinase.Virology. 2000 Feb 1;267(1):102-10. doi: 10.1006/viro.1999.0108. Virology. 2000. PMID: 10648187
Cited by
-
Infection of human cytomegalovirus in cultured human gingival tissue.Virol J. 2006 Oct 5;3:84. doi: 10.1186/1743-422X-3-84. Virol J. 2006. PMID: 17022821 Free PMC article.
-
Murine cytomegalovirus open reading frame M27 plays an important role in growth and virulence in mice.J Virol. 2001 Feb;75(4):1697-707. doi: 10.1128/JVI.75.4.1697-1707.2001. J Virol. 2001. PMID: 11160668 Free PMC article.
-
Murine gammaherpesvirus 68 open reading frame 31 is required for viral replication.J Virol. 2004 Jun;78(12):6610-20. doi: 10.1128/JVI.78.12.6610-6620.2004. J Virol. 2004. PMID: 15163752 Free PMC article.
-
Frequent coinfection of cells explains functional in vivo complementation between cytomegalovirus variants in the multiply infected host.J Virol. 2005 Aug;79(15):9492-502. doi: 10.1128/JVI.79.15.9492-9502.2005. J Virol. 2005. PMID: 16014912 Free PMC article.
-
Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene.J Virol. 2002 Mar;76(5):2316-28. doi: 10.1128/jvi.76.5.2316-2328.2002. J Virol. 2002. PMID: 11836410 Free PMC article.
References
-
- O'Connor M, Peifer M, Bender W. Science. 1989;244:1307–1312. - PubMed
-
- Horsburgh B C, Hubinette M M, Qiang D, MacDonald M L, Tufaro F. Gene Ther. 1999;6:922–930. - PubMed
-
- Saeki Y, Ichikawa T, Saeki A, Chiocca E A, Tobler K, Ackermann M, Breakefield X O, Fraefel C. Hum Gene Ther. 1998;9:2787–2794. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

