Role of magnesium chelatase activity in the early steps of the tetrapyrrole biosynthetic pathway
- PMID: 10759511
- PMCID: PMC58950
- DOI: 10.1104/pp.122.4.1161
Role of magnesium chelatase activity in the early steps of the tetrapyrrole biosynthetic pathway
Abstract
Magnesium-protoporphyrin IX chelatase (Mg-chelatase) is located at the branchpoint of tetrapyrrole biosynthesis, at which point protoporphyrin IX is distributed for the synthesis of chlorophyll and heme. We investigated the regulatory contribution of Mg-chelatase to the flow of metabolites. In plants, the enzyme complex consists of three subunits, designated CHL D, CHL I, and CHL H. Transgenic tobacco (Nicotiana tabacum) plants expressing antisense RNA for the Mg-chelatase subunit CHL H were analyzed to elucidate further the role of Mg-chelatase in the distribution of protoporphyrin IX into the branched tetrapyrrolic pathway. The transgenic plants displayed a reduced growth rate and chlorophyll deficiency. Both phenotypical properties were correlated with lower Mg-chelatase activity. Unexpectedly, less protoporphyrin IX and heme accumulated, and a decrease in 5-aminolevulinate (ALA)-synthesizing capacity and ALA dehydratase activity paralleled the progressive reduction in Mg-chelatase activity in the transformants compared with control plants. The reduced activities of the early enzymatic steps corresponded with lower levels of transcripts encoding glutamyl-tRNA reductase and ALA-dehydratase. The decreased expression and activities of early enzymes in the pathway could be explained by a feedback-controlled mechanism in response to lower Mg-chelatase activity. We discuss intercompartmental signaling that synchronizes the activities of the first steps in tetrapyrrolic metabolism with the late steps for the synthesis of end products.
Figures


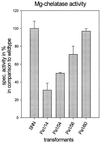
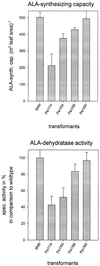

Similar articles
-
Decreased and increased expression of the subunit CHL I diminishes Mg chelatase activity and reduces chlorophyll synthesis in transgenic tobacco plants.Plant J. 2000 Apr;22(2):155-64. doi: 10.1046/j.1365-313x.2000.00724.x. Plant J. 2000. PMID: 10792831
-
Tobacco Mg protoporphyrin IX methyltransferase is involved in inverse activation of Mg porphyrin and protoheme synthesis.Plant J. 2005 Jan;41(2):282-90. doi: 10.1111/j.1365-313X.2004.02291.x. Plant J. 2005. PMID: 15634204
-
Metabolic control of the tetrapyrrole biosynthetic pathway for porphyrin distribution in the barley mutant albostrians.Plant J. 2003 Aug;35(4):512-22. doi: 10.1046/j.1365-313x.2003.01825.x. Plant J. 2003. PMID: 12904213
-
Novel insights in the control of tetrapyrrole metabolism of higher plants.Curr Opin Plant Biol. 1998 Jun;1(3):245-50. doi: 10.1016/s1369-5266(98)80112-x. Curr Opin Plant Biol. 1998. PMID: 10066589 Review.
-
Mechanism and regulation of Mg-chelatase.Biochem J. 1997 Oct 15;327 ( Pt 2)(Pt 2):321-33. doi: 10.1042/bj3270321. Biochem J. 1997. PMID: 9359397 Free PMC article. Review.
Cited by
-
Disturbance of chlorophyll formation at the level of 5-aminolevulinic acid and Mg-containing porphyrin synthesis in isogenic lines of spring wheat (Triticum aestivum L.) marked with genes cn-A1 and cn-D1.Dokl Biol Sci. 2005 Nov-Dec;405:472-3. doi: 10.1007/s10630-005-0169-8. Dokl Biol Sci. 2005. PMID: 16485648 No abstract available.
-
Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction.Proc Natl Acad Sci U S A. 2001 Feb 13;98(4):2053-8. doi: 10.1073/pnas.98.4.2053. Proc Natl Acad Sci U S A. 2001. PMID: 11172074 Free PMC article.
-
Chlorophyll biosynthesis under the control of arginine metabolism.Cell Rep. 2023 Nov 28;42(11):113265. doi: 10.1016/j.celrep.2023.113265. Epub 2023 Oct 20. Cell Rep. 2023. PMID: 37864789 Free PMC article.
-
Beyond identity: Understanding the contribution of the 5' nucleotide of the antisense strand to RNAi activity.PLoS One. 2021 Sep 7;16(9):e0256863. doi: 10.1371/journal.pone.0256863. eCollection 2021. PLoS One. 2021. PMID: 34492058 Free PMC article.
-
Transcriptome analysis based on a combination of sequencing platforms provides insights into leaf pigmentation in Acer rubrum.BMC Plant Biol. 2019 Jun 6;19(1):240. doi: 10.1186/s12870-019-1850-7. BMC Plant Biol. 2019. PMID: 31170934 Free PMC article.
References
-
- Beale SI, Weinstein JD. Tetrapyrrole metabolism in photosynthetic organisms. In: Dailey HA, editor. Biosynthesis of Heme and Chlorophylls. New York: McGraw-Hill; 1990. pp. 287–391.
-
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Castelfranco PA, Walker CJ, Weinstein JD. Biosynthetic studies on chlorophylls: from protoporphyrin IX to protochlorophyllide. In: Chadwick DJ, Ackrill K, editors. The Biosynthesis of the Tetrapyrrole Pigments. Ciba Foundation Symposium 180. Chichester, UK: John Wiley; 1994. pp. 194–209. - PubMed
-
- Chomczinsky P, Sacchi N. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

