Domains in the SPT5 protein that modulate its transcriptional regulatory properties
- PMID: 10757782
- PMCID: PMC85557
- DOI: 10.1128/MCB.20.9.2970-2983.2000
Domains in the SPT5 protein that modulate its transcriptional regulatory properties
Abstract
SPT5 and its binding partner SPT4 regulate transcriptional elongation by RNA polymerase II. SPT4 and SPT5 are involved in both 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB)-mediated transcriptional inhibition and the activation of transcriptional elongation by the human immunodeficiency virus type 1 (HIV-1) Tat protein. Recent data suggest that P-TEFb, which is composed of CDK9 and cyclin T1, is also critical in regulating transcriptional elongation by SPT4 and SPT5. In this study, we analyze the domains of SPT5 that regulate transcriptional elongation in the presence of either DRB or the HIV-1 Tat protein. We demonstrate that SPT5 domains that bind SPT4 and RNA polymerase II, in addition to a region in the C terminus of SPT5 that contains multiple heptad repeats and is designated CTR1, are critical for in vitro transcriptional repression by DRB and activation by the Tat protein. Furthermore, the SPT5 CTR1 domain is a substrate for P-TEFb phosphorylation. These results suggest that C-terminal repeats in SPT5, like those in the RNA polymerase II C-terminal domain, are sites for P-TEFb phosphorylation and function in modulating its transcriptional elongation properties.
Figures


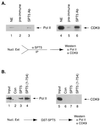
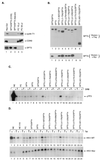



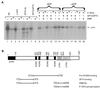
Similar articles
-
Phosphorylation of the RNA polymerase II carboxyl-terminal domain by CDK9 is directly responsible for human immunodeficiency virus type 1 Tat-activated transcriptional elongation.Mol Cell Biol. 2002 Jul;22(13):4622-37. doi: 10.1128/MCB.22.13.4622-4637.2002. Mol Cell Biol. 2002. PMID: 12052871 Free PMC article.
-
Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences.Mol Cell Biol. 2002 Feb;22(4):1079-93. doi: 10.1128/MCB.22.4.1079-1093.2002. Mol Cell Biol. 2002. PMID: 11809800 Free PMC article.
-
DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs.Genes Dev. 1998 Feb 1;12(3):343-56. doi: 10.1101/gad.12.3.343. Genes Dev. 1998. PMID: 9450929 Free PMC article.
-
Transcription elongation: the 'Foggy' is liftingellipsis.Curr Biol. 2001 Feb 20;11(4):R144-6. doi: 10.1016/s0960-9822(01)00063-x. Curr Biol. 2001. PMID: 11250170 Review.
-
The Spt4-Spt5 complex: a multi-faceted regulator of transcription elongation.Biochim Biophys Acta. 2013 Jan;1829(1):105-15. doi: 10.1016/j.bbagrm.2012.08.007. Epub 2012 Sep 6. Biochim Biophys Acta. 2013. PMID: 22982195 Free PMC article. Review.
Cited by
-
Control of HIV latency by epigenetic and non-epigenetic mechanisms.Curr HIV Res. 2011 Dec 1;9(8):554-67. doi: 10.2174/157016211798998736. Curr HIV Res. 2011. PMID: 22211660 Free PMC article.
-
Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, seliciclib and flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex.Retrovirology. 2007 Jul 11;4:47. doi: 10.1186/1742-4690-4-47. Retrovirology. 2007. PMID: 17625008 Free PMC article.
-
P-TEFb is degraded by Siah1/2 in quiescent cells.Nucleic Acids Res. 2022 May 20;50(9):5000-5013. doi: 10.1093/nar/gkac291. Nucleic Acids Res. 2022. PMID: 35524561 Free PMC article.
-
A new paradigm in eukaryotic biology: HIV Tat and the control of transcriptional elongation.PLoS Biol. 2005 Feb;3(2):e76. doi: 10.1371/journal.pbio.0030076. PLoS Biol. 2005. PMID: 15719065 Free PMC article.
-
The positive transcription elongation factor b is an essential cofactor for the activation of transcription by myocyte enhancer factor 2.J Mol Biol. 2008 Oct 3;382(2):275-87. doi: 10.1016/j.jmb.2008.07.017. Epub 2008 Jul 16. J Mol Biol. 2008. PMID: 18662700 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
