Development of reverse transcription-PCR assays specific for detection of equine encephalitis viruses
- PMID: 10747138
- PMCID: PMC86482
- DOI: 10.1128/JCM.38.4.1527-1535.2000
Development of reverse transcription-PCR assays specific for detection of equine encephalitis viruses
Abstract
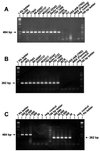
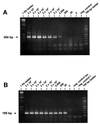
Specific and sensitive reverse transcription-PCR (RT-PCR) assays were developed for the detection of eastern, western, and Venezuelan equine encephalitis viruses (EEE, WEE, and VEE, respectively). Tests for specificity included all known alphavirus species. The EEE-specific RT-PCR amplified a 464-bp region of the E2 gene exclusively from 10 different EEE strains from South and North America with a sensitivity of about 3,000 RNA molecules. In a subsequent nested PCR, the specificity was confirmed by the amplification of a 262-bp fragment, increasing the sensitivity of this assay to approximately 30 RNA molecules. The RT-PCR for WEE amplified a fragment of 354 bp from as few as 2,000 RNA molecules. Babanki virus, as well as Mucambo and Pixuna viruses (VEE subtypes IIIA and IV), were also amplified. However, the latter viruses showed slightly smaller fragments of about 290 and 310 bp, respectively. A subsequent seminested PCR amplified a 195-bp fragment only from the 10 tested strains of WEE from North and South America, rendering this assay virus specific and increasing its sensitivity to approximately 20 RNA molecules. Because the 12 VEE subtypes showed too much divergence in their 26S RNA nucleotide sequences to detect all of them by the use of nondegenerate primers, this assay was confined to the medically important and closely related VEE subtypes IAB, IC, ID, IE, and II. The RT-PCR-seminested PCR combination specifically amplified 342- and 194-bp fragments of the region covering the 6K gene in VEE. The sensitivity was 20 RNA molecules for subtype IAB virus and 70 RNA molecules for subtype IE virus. In addition to the subtypes mentioned above, three of the enzootic VEE (subtypes IIIB, IIIC, and IV) showed the specific amplicon in the seminested PCR. The practicability of the latter assay was tested with human sera gathered as part of the febrile illness surveillance in the Amazon River Basin of Peru near the city of Iquitos. All of the nine tested VEE-positive sera showed the expected 194-bp amplicon of the VEE-specific RT-PCR-seminested PCR.
Figures



Similar articles
-
A duplex real-time reverse transcriptase polymerase chain reaction assay for detecting western equine and eastern equine encephalitis viruses.Virol J. 2010 Oct 26;7:284. doi: 10.1186/1743-422X-7-284. Virol J. 2010. PMID: 20977706 Free PMC article.
-
Cross-protective immunity between equine encephalomyelitis viruses in equids.Am J Vet Res. 1989 Sep;50(9):1442-6. Am J Vet Res. 1989. PMID: 2552875
-
Detection of North American eastern and western equine encephalitis viruses by nucleic acid amplification assays.J Clin Microbiol. 2003 Jan;41(1):379-85. doi: 10.1128/JCM.41.1.379-385.2003. J Clin Microbiol. 2003. PMID: 12517876 Free PMC article.
-
[EPIDEMIOLOGIC ANALYSIS OF OUTBREAKS OF DISEASES CAUSED BY AMERICAN EQUINE ENCEPHALITIS CAUSATIVE AGENTS IN ENDEMIC REGIONS].Zh Mikrobiol Epidemiol Immunobiol. 2015 Sep-Oct;(5):103-10. Zh Mikrobiol Epidemiol Immunobiol. 2015. PMID: 26829861 Review. Russian.
-
Alphaviral equine encephalomyelitis (Eastern, Western and Venezuelan).Rev Sci Tech. 2015 Aug;34(2):491-501. doi: 10.20506/rst.34.2.2374. Rev Sci Tech. 2015. PMID: 26601451 Review.
Cited by
-
Detection of viruses in used ventilation filters from two large public buildings.Am J Infect Control. 2011 Sep;39(7):e30-8. doi: 10.1016/j.ajic.2010.10.036. Epub 2011 May 6. Am J Infect Control. 2011. PMID: 21549446 Free PMC article.
-
A duplex real-time reverse transcriptase polymerase chain reaction assay for detecting western equine and eastern equine encephalitis viruses.Virol J. 2010 Oct 26;7:284. doi: 10.1186/1743-422X-7-284. Virol J. 2010. PMID: 20977706 Free PMC article.
-
Real-time RT-PCR for Venezuelan equine encephalitis complex, Madariaga, and Eastern equine encephalitis viruses: application in human and mosquito public health surveillance in Panama.J Clin Microbiol. 2023 Dec 19;61(12):e0015223. doi: 10.1128/jcm.00152-23. Epub 2023 Nov 20. J Clin Microbiol. 2023. PMID: 37982611 Free PMC article.
-
A duplex real-time reverse transcriptase polymerase chain reaction assay for the detection of St. Louis encephalitis and eastern equine encephalitis viruses.Diagn Microbiol Infect Dis. 2008 Nov;62(3):272-9. doi: 10.1016/j.diagmicrobio.2008.07.004. Epub 2008 Aug 20. Diagn Microbiol Infect Dis. 2008. PMID: 18715737 Free PMC article.
-
Rapid identification of emerging infectious agents using PCR and electrospray ionization mass spectrometry.Ann N Y Acad Sci. 2007 Apr;1102(1):109-20. doi: 10.1196/annals.1408.008. Ann N Y Acad Sci. 2007. PMID: 17470915 Free PMC article. Review.
References
-
- Armstrong P, Borovsky D, Shope R E, Morris C D, Mitchell C J, Karabatsos N, Komar N, Spielman A. Sensitive and specific colorimetric dot assay to detect eastern equine encephalomyelitis viral RNA in mosquitos (Diptera: Culicidae) after polymerase chain reaction amplification. J Med Entomol. 1995;32:42–52. - PubMed
-
- Brightwell G, Brown J M, Coates D M. Genetic targets for the detection and identification of Venezuelan equine encephalitis viruses. Arch Virol. 1998;143:731–742. - PubMed
-
- Calisher C H, Karabatsos N, Lazuick J S, Monath T P, Wolff K L. Reevaluation of the western equine encephalitis antigenic complex of alphaviruses (family Togaviridae) as determined by neutralization tests. Am J Trop Med Hyg. 1988;38:447–452. - PubMed
-
- Calisher C H, Kinney R M, de Souza Lopes O, Trent D W, Monath T P, France D B. Identification of a new Venezuelan equine encephalitis virus from Brazil. Am J Trop Med Hyg. 1982;31:1260–1272. - PubMed
-
- Calisher C H, Monath T P, Sabattini M S, Cropp C B, Kerschner J, Hunt A R, Lazuick J S. Arbovirus investigations in Argentinia, 1977–1980. III. Identification and characterization of viruses isolated, including new subtypes of western and Venezuelan equine encephalitis viruses and four new bunyaviruses (Las Malayos, Resistencia, Barranqueras, and Antequera) Am J Trop Med Hyg. 1985;34:956–965. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

