Active and alkylated human AGT structures: a novel zinc site, inhibitor and extrahelical base binding
- PMID: 10747039
- PMCID: PMC310240
- DOI: 10.1093/emboj/19.7.1719
Active and alkylated human AGT structures: a novel zinc site, inhibitor and extrahelical base binding
Abstract
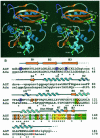
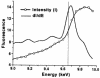
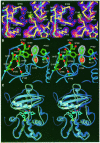
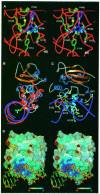
Human O(6)-alkylguanine-DNA alkyltransferase (AGT), which directly reverses endogenous alkylation at the O(6)-position of guanine, confers resistance to alkylation chemotherapies and is therefore an active anticancer drug target. Crystal structures of active human AGT and its biologically and therapeutically relevant methylated and benzylated product complexes reveal an unexpected zinc-stabilized helical bridge joining a two-domain alpha/beta structure. An asparagine hinge couples the active site motif to a helix-turn-helix (HTH) motif implicated in DNA binding. The reactive cysteine environment, its position within a groove adjacent to the alkyl-binding cavity and mutational analyses characterize DNA-damage recognition and inhibitor specificity, support a structure-based dealkylation mechanism and suggest a molecular basis for destabilization of the alkylated protein. These results support damaged nucleotide flipping facilitated by an arginine finger within the HTH motif to stabilize the extrahelical O(6)-alkylguanine without the protein conformational change originally proposed from the empty Ada structure. Cysteine alkylation sterically shifts the HTH recognition helix to evidently mechanistically couple release of repaired DNA to an opening of the protein fold to promote the biological turnover of the alkylated protein.
Figures
Similar articles
-
Conserved structural motifs governing the stoichiometric repair of alkylated DNA by O(6)-alkylguanine-DNA alkyltransferase.Mutat Res. 2000 Aug 30;460(3-4):151-63. doi: 10.1016/s0921-8777(00)00024-0. Mutat Res. 2000. PMID: 10946226 Review.
-
DNA binding, nucleotide flipping, and the helix-turn-helix motif in base repair by O6-alkylguanine-DNA alkyltransferase and its implications for cancer chemotherapy.DNA Repair (Amst). 2007 Aug 1;6(8):1100-15. doi: 10.1016/j.dnarep.2007.03.011. Epub 2007 May 7. DNA Repair (Amst). 2007. PMID: 17485252 Free PMC article. Review.
-
DNA binding and nucleotide flipping by the human DNA repair protein AGT.Nat Struct Mol Biol. 2004 Aug;11(8):714-20. doi: 10.1038/nsmb791. Epub 2004 Jun 27. Nat Struct Mol Biol. 2004. PMID: 15221026
-
Investigation of the role of tyrosine-114 in the activity of human O6-alkylguanine-DNA alkyltranferase.Biochemistry. 1998 Sep 8;37(36):12489-95. doi: 10.1021/bi9811718. Biochemistry. 1998. PMID: 9730821
-
Active-site alkylation destabilizes human O6-alkylguanine DNA alkyltransferase.Protein Sci. 2004 Jan;13(1):301-5. doi: 10.1110/ps.03319404. Protein Sci. 2004. PMID: 14691244 Free PMC article.
Cited by
-
S-nitrosylation from GSNOR deficiency impairs DNA repair and promotes hepatocarcinogenesis.Sci Transl Med. 2010 Feb 17;2(19):19ra13. doi: 10.1126/scitranslmed.3000328. Sci Transl Med. 2010. PMID: 20371487 Free PMC article.
-
DNA-binding mechanism of the Escherichia coli Ada O(6)-alkylguanine-DNA alkyltransferase.Nucleic Acids Res. 2000 Oct 1;28(19):3710-8. doi: 10.1093/nar/28.19.3710. Nucleic Acids Res. 2000. PMID: 11000262 Free PMC article.
-
AdoMet-dependent methylation, DNA methyltransferases and base flipping.Nucleic Acids Res. 2001 Sep 15;29(18):3784-95. doi: 10.1093/nar/29.18.3784. Nucleic Acids Res. 2001. PMID: 11557810 Free PMC article. Review.
-
The modified human DNA repair enzyme O(6)-methylguanine-DNA methyltransferase is a negative regulator of estrogen receptor-mediated transcription upon alkylation DNA damage.Mol Cell Biol. 2001 Oct;21(20):7105-14. doi: 10.1128/MCB.21.20.7105-7114.2001. Mol Cell Biol. 2001. PMID: 11564893 Free PMC article.
-
Alkylation damage repair protein O6-alkylguanine-DNA alkyltransferase from the hyperthermophiles Aquifex aeolicus and Archaeoglobus fulgidus.Biochem J. 2003 Oct 15;375(Pt 2):449-55. doi: 10.1042/BJ20030809. Biochem J. 2003. PMID: 12892560 Free PMC article.
References
-
- Beranek D.T. (1990) Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat. Res., 231, 11–30. - PubMed
-
- Brennan R.G. (1993) The winged–helix DNA-binding motif: another helix–turn–helix takeoff. Cell, 74, 773–776. - PubMed
-
- Brunger A.T., et al. (1998)Crystallography and NMR system (CNS): A new software system for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. - PubMed
-
- Casadevall M. and Sarkar, B. (1998) Effect of redox conditions on the DNA-binding efficiency of the retinoic acid receptor zinc-finger. J. Inorg. Biochem., 71, 147–152. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

