Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion
- PMID: 10737792
- PMCID: PMC16226
- DOI: 10.1073/pnas.97.7.3260
Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion
Abstract
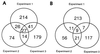
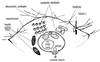
MYC affects normal and neoplastic cell proliferation by altering gene expression, but the precise pathways remain unclear. We used oligonucleotide microarray analysis of 6,416 genes and expressed sequence tags to determine changes in gene expression caused by activation of c-MYC in primary human fibroblasts. In these experiments, 27 genes were consistently induced, and 9 genes were repressed. The identity of the genes revealed that MYC may affect many aspects of cell physiology altered in transformed cells: cell growth, cell cycle, adhesion, and cytoskeletal organization. Identified targets possibly linked to MYC's effects on cell growth include the nucleolar proteins nucleolin and fibrillarin, as well as the eukaryotic initiation factor 5A. Among the cell cycle genes identified as targets, the G1 cyclin D2 and the cyclin-dependent kinase binding protein CksHs2 were induced whereas the cyclin-dependent kinase inhibitor p21(Cip1) was repressed. A role for MYC in regulating cell adhesion and structure is suggested by repression of genes encoding the extracellular matrix proteins fibronectin and collagen, and the cytoskeletal protein tropomyosin. A possible mechanism for MYC-mediated apoptosis was revealed by identification of the tumor necrosis factor receptor associated protein TRAP1 as a MYC target. Finally, two immunophilins, peptidyl-prolyl cis-trans isomerase F and FKBP52, the latter of which plays a role in cell division in Arabidopsis, were up-regulated by MYC. We also explored pattern-matching methods as an alternative approach for identifying MYC target genes. The genes that displayed an expression profile most similar to endogenous Myc in microarray-based expression profiling of myeloid differentiation models were highly enriched for MYC target genes.
Figures
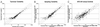
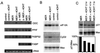
Similar articles
-
Adhesion-regulated G1 cell cycle arrest in epithelial cells requires the downregulation of c-Myc.Oncogene. 2001 Jul 27;20(33):4554-67. doi: 10.1038/sj.onc.1204609. Oncogene. 2001. PMID: 11494151
-
Regulation of cell cycle entry and G1 progression by CSF-1.Mol Reprod Dev. 1997 Jan;46(1):11-8. doi: 10.1002/(SICI)1098-2795(199701)46:1<11::AID-MRD3>3.0.CO;2-U. Mol Reprod Dev. 1997. PMID: 8981358 Review.
-
Mechanisms of growth arrest by c-myc antisense oligonucleotides in MCF-7 breast cancer cells: implications for the antiproliferative effects of antiestrogens.Cancer Res. 2002 Jun 1;62(11):3126-31. Cancer Res. 2002. PMID: 12036924
-
A modest reduction in c-myc expression has minimal effects on cell growth and apoptosis but dramatically reduces susceptibility to Ras and Raf transformation.Cancer Res. 2001 Feb 1;61(3):1178-86. Cancer Res. 2001. PMID: 11221849
-
The proto-oncogene c-myc in hematopoietic development and leukemogenesis.Oncogene. 2002 May 13;21(21):3414-21. doi: 10.1038/sj.onc.1205400. Oncogene. 2002. PMID: 12032779 Review.
Cited by
-
The translation factor eIF5A and human cancer.Biochim Biophys Acta. 2015 Jul;1849(7):836-44. doi: 10.1016/j.bbagrm.2015.05.002. Epub 2015 May 13. Biochim Biophys Acta. 2015. PMID: 25979826 Free PMC article. Review.
-
miR-1204 Positioning in 8q24.21 Involved in the Tumorigenesis of Colorectal Cancer by Targeting MASPIN.Protein Pept Lett. 2024;31(7):544-558. doi: 10.2174/0109298665305114240718072029. Protein Pept Lett. 2024. PMID: 39082173
-
Single-cell RNA-sequencing identifies the developmental trajectory of C-Myc-dependent NK1.1- T-bet+ intraepithelial lymphocyte precursors.Mucosal Immunol. 2020 Mar;13(2):257-270. doi: 10.1038/s41385-019-0220-y. Epub 2019 Nov 11. Mucosal Immunol. 2020. PMID: 31712600 Free PMC article.
-
A core MYC gene expression signature is prominent in basal-like breast cancer but only partially overlaps the core serum response.PLoS One. 2009 Aug 19;4(8):e6693. doi: 10.1371/journal.pone.0006693. PLoS One. 2009. PMID: 19690609 Free PMC article.
-
The limit fold change model: a practical approach for selecting differentially expressed genes from microarray data.BMC Bioinformatics. 2002 Jun 21;3:17. doi: 10.1186/1471-2105-3-17. BMC Bioinformatics. 2002. PMID: 12095422 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

