Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression
- PMID: 10715325
- PMCID: PMC139839
- DOI: 10.1105/tpc.12.3.393
Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression
Abstract
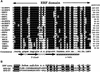
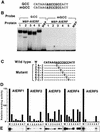
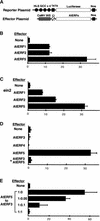
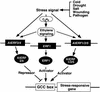
Ethylene-responsive element binding factors (ERFs) are members of a novel family of transcription factors that are specific to plants. A highly conserved DNA binding domain known as the ERF domain is the unique feature of this protein family. To characterize in detail this family of transcription factors, we isolated Arabidopsis cDNAs encoding five different ERF proteins (AtERF1 to AtERF5) and analyzed their structure, DNA binding preference, transactivation ability, and mRNA expression profiles. The isolated AtERFs were placed into three classes based on amino acid identity within the ERF domain, although all five displayed GCC box-specific binding activity. AtERF1, AtERF2, and AtERF5 functioned as activators of GCC box-dependent transcription in Arabidopsis leaves. By contrast, AtERF3 and AtERF4 acted as repressors that downregulated not only basal transcription levels of a reporter gene but also the transactivation activity of other transcription factors. The AtERF genes were differentially regulated by ethylene and by abiotic stress conditions, such as wounding, cold, high salinity, or drought, via ETHYLENE-INSENSITIVE2 (EIN2)-dependent or -independent pathways. Cycloheximide, a protein synthesis inhibitor, also induced marked accumulation of AtERF mRNAs. Thus, we conclude that AtERFs are factors that respond to extracellular signals to modulate GCC box-mediated gene expression positively or negatively.
Figures




Similar articles
-
The ethylene-responsive factor like protein 1 (CaERFLP1) of hot pepper (Capsicum annuum L.) interacts in vitro with both GCC and DRE/CRT sequences with different binding affinities: possible biological roles of CaERFLP1 in response to pathogen infection and high salinity conditions in transgenic tobacco plants.Plant Mol Biol. 2004 May;55(1):61-81. doi: 10.1007/s11103-004-0417-6. Plant Mol Biol. 2004. PMID: 15604665
-
Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses.Plant Mol Biol. 2005 Jul;58(4):585-96. doi: 10.1007/s11103-005-7294-5. Plant Mol Biol. 2005. PMID: 16021341
-
Molecular dynamics simulations reveal the disparity in specific recognition of GCC-box by AtERFs transcription factors super family in Arabidopsis.J Mol Recognit. 2009 Nov-Dec;22(6):474-9. doi: 10.1002/jmr.965. J Mol Recognit. 2009. PMID: 19533627
-
Regulation of ethylene-induced transcription of defense genes.Plant Cell Physiol. 2000 Nov;41(11):1187-92. doi: 10.1093/pcp/pcd057. Plant Cell Physiol. 2000. PMID: 11092902 Review.
-
Emerging role of the plant ERF transcription factors in coordinating wound defense responses and repair.J Cell Sci. 2018 Jan 29;131(2):jcs208215. doi: 10.1242/jcs.208215. J Cell Sci. 2018. PMID: 29242229 Review.
Cited by
-
Discovery of core biotic stress responsive genes in Arabidopsis by weighted gene co-expression network analysis.PLoS One. 2015 Mar 2;10(3):e0118731. doi: 10.1371/journal.pone.0118731. eCollection 2015. PLoS One. 2015. PMID: 25730421 Free PMC article.
-
The ARGOS gene family functions in a negative feedback loop to desensitize plants to ethylene.BMC Plant Biol. 2015 Jun 24;15:157. doi: 10.1186/s12870-015-0554-x. BMC Plant Biol. 2015. PMID: 26105742 Free PMC article.
-
Divergent DNA-binding specificities of a group of ETHYLENE RESPONSE FACTOR transcription factors involved in plant defense.Plant Physiol. 2013 Jun;162(2):977-90. doi: 10.1104/pp.113.217455. Epub 2013 Apr 29. Plant Physiol. 2013. PMID: 23629834 Free PMC article.
-
Characterization of a novel ERF transcription factor in Artemisia annua and its induction kinetics after hormones and stress treatments.Mol Biol Rep. 2012 Oct;39(10):9521-7. doi: 10.1007/s11033-012-1816-4. Epub 2012 Jun 20. Mol Biol Rep. 2012. PMID: 22714923
-
Phytohormones as Regulators of Mitochondrial Gene Expression in Arabidopsis thaliana.Int J Mol Sci. 2023 Nov 29;24(23):16924. doi: 10.3390/ijms242316924. Int J Mol Sci. 2023. PMID: 38069246 Free PMC article.
References
-
- Abeles, F.B., Morgan, P.W., and Saltveit, M.E., Jr. (1992). Ethylene in Plant Biology, 2nd ed. (San Diego, CA: Academic Press).
-
- Berberich, T., and Kusano, T. (1997). Cycloheximide induces a subset of low temperature–inducible genes in maize. Mol. Gen. Genet. 254 275–283. - PubMed
-
- Chandler, P.M., and Robertson, M. (1994). Gene expression regulated by abscisic acid and its relation to stress tolerance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45 113–141.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

