DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA
- PMID: 10675345
- PMCID: PMC305614
- DOI: 10.1093/emboj/19.4.758
DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA
Abstract
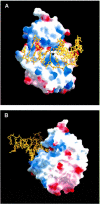
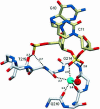
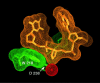
The Escherichia coli AlkA protein is a base excision repair glycosylase that removes a variety of alkylated bases from DNA. The 2.5 A crystal structure of AlkA complexed to DNA shows a large distortion in the bound DNA. The enzyme flips a 1-azaribose abasic nucleotide out of DNA and induces a 66 degrees bend in the DNA with a marked widening of the minor groove. The position of the 1-azaribose in the enzyme active site suggests an S(N)1-type mechanism for the glycosylase reaction, in which the essential catalytic Asp238 provides direct assistance for base removal. Catalytic selectivity might result from the enhanced stacking of positively charged, alkylated bases against the aromatic side chain of Trp272 in conjunction with the relative ease of cleaving the weakened glycosylic bond of these modified nucleotides. The structure of the AlkA-DNA complex offers the first glimpse of a helix-hairpin-helix (HhH) glycosylase complexed to DNA. Modeling studies suggest that other HhH glycosylases can bind to DNA in a similar manner.
Figures
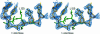
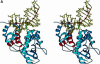
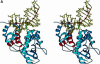
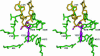

Similar articles
-
Structural bases for substrate recognition and repair system of base-excision DNA repair proteins.Nucleic Acids Symp Ser. 2000;(44):57-8. doi: 10.1093/nass/44.1.57. Nucleic Acids Symp Ser. 2000. PMID: 12903266
-
Crystal structures of human DNA polymerase beta complexed with DNA: implications for catalytic mechanism, processivity, and fidelity.Biochemistry. 1996 Oct 1;35(39):12742-61. doi: 10.1021/bi952955d. Biochemistry. 1996. PMID: 8841118
-
Structural studies of human alkyladenine glycosylase and E. coli 3-methyladenine glycosylase.Mutat Res. 2000 Aug 30;460(3-4):201-10. doi: 10.1016/s0921-8777(00)00027-6. Mutat Res. 2000. PMID: 10946229 Review.
-
A novel 3-methyladenine DNA glycosylase from Helicobacter pylori defines a new class within the endonuclease III family of base excision repair glycosylases.J Biol Chem. 2000 Jun 30;275(26):20077-83. doi: 10.1074/jbc.M001071200. J Biol Chem. 2000. PMID: 10777493
-
Three-dimensional structural views of damaged-DNA recognition: T4 endonuclease V, E. coli Vsr protein, and human nucleotide excision repair factor XPA.Mutat Res. 2000 Aug 30;460(3-4):257-75. doi: 10.1016/s0921-8777(00)00031-8. Mutat Res. 2000. PMID: 10946233 Review.
Cited by
-
Insights into the substrate discrimination mechanisms of methyl-CpG-binding domain 4.Biochem J. 2021 May 28;478(10):1985-1997. doi: 10.1042/BCJ20210017. Biochem J. 2021. PMID: 33960375 Free PMC article.
-
Recent advances in the structural mechanisms of DNA glycosylases.Biochim Biophys Acta. 2013 Jan;1834(1):247-71. doi: 10.1016/j.bbapap.2012.10.005. Epub 2012 Oct 14. Biochim Biophys Acta. 2013. PMID: 23076011 Free PMC article. Review.
-
Sculpting of DNA at abasic sites by DNA glycosylase homolog mag2.Structure. 2013 Jan 8;21(1):154-166. doi: 10.1016/j.str.2012.11.004. Epub 2012 Dec 13. Structure. 2013. PMID: 23245849 Free PMC article.
-
Duplex interrogation by a direct DNA repair protein in search of base damage.Nat Struct Mol Biol. 2012 Jun 3;19(7):671-6. doi: 10.1038/nsmb.2320. Nat Struct Mol Biol. 2012. PMID: 22659876 Free PMC article.
-
Crystal structure of human methyl-binding domain IV glycosylase bound to abasic DNA.J Mol Biol. 2012 Jul 13;420(3):164-75. doi: 10.1016/j.jmb.2012.04.028. Epub 2012 May 2. J Mol Biol. 2012. PMID: 22560993 Free PMC article.
References
-
- Barrett T.E., Savva, R., Panayotou, G., Barlow, T., Brown, T., Jiricny, J. and Pearl, L.H. (1998) Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. Cell, 92, 117–129. - PubMed
-
- Brünger A.T. (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature, 355, 472–475. - PubMed
-
- Brünger A.T., et al. (1998)Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. - PubMed
-
- Deng L., Scharer, O.D. and Verdine, G.L. (1997) Unusually strong binding of a designed transition-state analog to a base-excision DNA repair protein. J. Am. Chem. Soc., 119, 7865–7866.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

