Regulation of E2F1 activity by acetylation
- PMID: 10675335
- PMCID: PMC305604
- DOI: 10.1093/emboj/19.4.662
Regulation of E2F1 activity by acetylation
Abstract
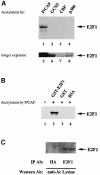
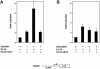
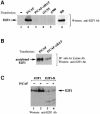
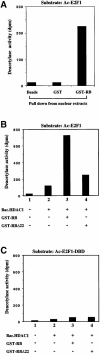
During the G(1) phase of the cell cycle, an E2F-RB complex represses transcription, via the recruitment of histone deacetylase activity. Phosphorylation of RB at the G(1)/S boundary generates a pool of 'free' E2F, which then stimulates transcription of S-phase genes. Given that E2F1 activity is stimulated by p300/CBP acetylase and repressed by an RB-associated deacetylase, we asked if E2F1 was subject to modification by acetylation. We show that the p300/CBP-associated factor P/CAF, and to a lesser extent p300/CBP itself, can acetylate E2F1 in vitro and that intracellular E2F1 is acetylated. The acetylation sites lie adjacent to the E2F1 DNA-binding domain and involve lysine residues highly conserved in E2F1, 2 and 3. Acetylation by P/CAF has three functional consequences on E2F1 activity: increased DNA-binding ability, activation potential and protein half-life. These results suggest that acetylation stimulates the functions of the non-RB bound 'free' form of E2F1. Consistent with this, we find that the RB-associated histone deacetylase can deacetylate E2F1. These results identify acetylation as a novel regulatory modification that stimulates E2F1's activation functions.
Figures




Similar articles
-
CBP/p300 histone acetyl-transferase activity is important for the G1/S transition.Oncogene. 2000 May 11;19(20):2430-7. doi: 10.1038/sj.onc.1203562. Oncogene. 2000. PMID: 10828885
-
Retinoblastoma protein represses transcription by recruiting a histone deacetylase.Nature. 1998 Feb 5;391(6667):601-5. doi: 10.1038/35410. Nature. 1998. PMID: 9468140
-
Retinoblastoma protein recruits histone deacetylase to repress transcription.Nature. 1998 Feb 5;391(6667):597-601. doi: 10.1038/35404. Nature. 1998. PMID: 9468139
-
Cell cycle: Flies teach an old dogma new tricks.Curr Biol. 2001 Mar 6;11(5):R178-81. doi: 10.1016/s0960-9822(01)00088-4. Curr Biol. 2001. PMID: 11267887 Review.
-
Introduction to the E2F family: protein structure and gene regulation.Curr Top Microbiol Immunol. 1996;208:1-30. doi: 10.1007/978-3-642-79910-5_1. Curr Top Microbiol Immunol. 1996. PMID: 8575210 Review.
Cited by
-
Keratins in colorectal epithelial function and disease.Int J Exp Pathol. 2012 Oct;93(5):305-18. doi: 10.1111/j.1365-2613.2012.00830.x. Int J Exp Pathol. 2012. PMID: 22974212 Free PMC article. Review.
-
Critical review of non-histone human substrates of metal-dependent lysine deacetylases.FASEB J. 2020 Oct;34(10):13140-13155. doi: 10.1096/fj.202001301RR. Epub 2020 Aug 30. FASEB J. 2020. PMID: 32862458 Free PMC article. Review.
-
Regulation of inositol 1,3,4-trisphosphate 5/6-kinase (ITPK1) by reversible lysine acetylation.Proc Natl Acad Sci U S A. 2012 Feb 14;109(7):2290-5. doi: 10.1073/pnas.1119740109. Epub 2012 Jan 30. Proc Natl Acad Sci U S A. 2012. PMID: 22308441 Free PMC article.
-
The histone acetyltransferases CBP/p300 are degraded in NIH 3T3 cells by activation of Ras signalling pathway.Biochem J. 2006 Sep 1;398(2):215-24. doi: 10.1042/BJ20060052. Biochem J. 2006. PMID: 16704373 Free PMC article.
-
Interaction between acetylated MyoD and the bromodomain of CBP and/or p300.Mol Cell Biol. 2001 Aug;21(16):5312-20. doi: 10.1128/MCB.21.16.5312-5320.2001. Mol Cell Biol. 2001. PMID: 11463815 Free PMC article.
References
-
- Allen K.E., de la Luna, S., Kerkhoven, R.M., Bernards, R. and La Thangue, N.B. (1997) Distinct mechanisms of nuclear accumulation regulate the functional consequence of E2F transcription factors. J. Cell Sci., 110, 2819–2831. - PubMed
-
- Bagchi S., Weinmann, R. and Raychaudhuri, P. (1991) The retinoblastoma protein copurifies with E2F-I, an E1A-regulated inhibitor of the transcription factor E2F. Cell, 65, 1063–1072. - PubMed
-
- Bannister A.J. and Kouzarides, T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. - PubMed
-
- Boyes J., Byfield, P., Nakatani, Y. and Ogryzko, V. (1998) Regulation of activity of the transcription factor GATA-1 by acetylation. Nature, 396, 594–598. - PubMed
-
- Brehm A., Miska, E.A., McCance, D.J., Reid, J.L., Bannister, A.J. and Kouzarides, T. (1998) Retinoblastoma protein recruits deacetylase to repress transcription. Nature, 391, 597–601. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

