Development of a real-time quantitative assay for detection of Epstein-Barr virus
- PMID: 10655372
- PMCID: PMC86184
- DOI: 10.1128/JCM.38.2.712-715.2000
Development of a real-time quantitative assay for detection of Epstein-Barr virus
Abstract
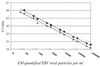
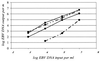
With the use of real-time PCR, we developed and evaluated a rapid, sensitive, specific, and reproducible method for the detection of Epstein-Barr virus (EBV) DNA in plasma samples. This method allowed us to screen plasma and serum samples over a range between 100 and 10(7) copies of DNA per ml using two sample preparation methods based on absorption. A precision study yielded an average coefficient of variation for both methods of less than 12%, with a coefficient of regression for the standard curve of a minimum of 0. 98. We detected EBV DNA in 19.2% of plasma samples from immunosuppressed solid-organ transplant patients without symptoms of EBV infections with a mean load of 440 copies per ml. EBV DNA could be detected in all transplant patients diagnosed with posttransplant lymphoproliferative disorder, with a mean load of 544,570 copies per ml. No EBV DNA could be detected in healthy individuals in nonimmunosuppressed control groups and a mean of 6,400 copies per ml could be detected in patients with infectious mononucleosis. Further studies revealed that the inhibitory effect of heparinized plasma could be efficiently removed by use of an extraction method with Celite as the absorbent.
Figures
Similar articles
-
Epstein Barr viral load monitoring by quantitative PCR in renal transplant patients.New Microbiol. 2003 Apr;26(2):141-9. New Microbiol. 2003. PMID: 12737195
-
Assessment of automated DNA extraction coupled with real-time PCR for measuring Epstein-Barr virus load in whole blood, peripheral mononuclear cells and plasma.J Clin Virol. 2004 Jun;30(2):157-64. doi: 10.1016/j.jcv.2003.10.002. J Clin Virol. 2004. PMID: 15125872
-
Viral DNA Loads in Various Blood Components of Patients With Epstein-Barr Virus-Positive T-Cell/Natural Killer Cell Lymphoproliferative Diseases.J Infect Dis. 2019 Sep 13;220(8):1307-1311. doi: 10.1093/infdis/jiz315. J Infect Dis. 2019. PMID: 31240305
-
Diagnostic testing in Epstein-Barr virus infection.Clin Chem Lab Med. 2001 Sep;39(9):789-94. doi: 10.1515/CCLM.2001.130. Clin Chem Lab Med. 2001. PMID: 11601674 Review.
-
Role of Epstein-Barr virus DNA load monitoring in prevention and early detection of post-transplant lymphoproliferative disease.Leuk Lymphoma. 2002 Apr;43(4):831-40. doi: 10.1080/10428190290016971. Leuk Lymphoma. 2002. PMID: 12153173 Review.
Cited by
-
Failure to detect Epstein-Barr virus (EBV) DNA in plasma by real-time PCR in a case of EBV-associated posttransplantation lymphoproliferative disorder confined to the central nervous system.Int J Hematol. 2002 May;75(4):416-20. doi: 10.1007/BF02982135. Int J Hematol. 2002. PMID: 12041675
-
The role of the virology laboratory in the management of hepatitis C virus infection.J Clin Virol. 2002 Jul;25(1):3-13. doi: 10.1016/s1386-6532(02)00026-4. J Clin Virol. 2002. PMID: 12126716 Free PMC article. Review.
-
Advances in real-time PCR: application to clinical laboratory diagnostics.Adv Clin Chem. 2005;40:219-59. doi: 10.1016/s0065-2423(05)40006-2. Adv Clin Chem. 2005. PMID: 16355924 Free PMC article. Review.
-
Comparison of various blood compartments and reporting units for the detection and quantification of Epstein-Barr virus in peripheral blood.J Clin Microbiol. 2007 Jul;45(7):2151-5. doi: 10.1128/JCM.02308-06. Epub 2007 May 9. J Clin Microbiol. 2007. PMID: 17494720 Free PMC article.
-
Epstein-barr virus coinfection in cerebrospinal fluid is associated with increased mortality in Malawian adults with bacterial meningitis.J Infect Dis. 2012 Jan 1;205(1):106-10. doi: 10.1093/infdis/jir707. Epub 2011 Nov 9. J Infect Dis. 2012. PMID: 22075766 Free PMC article.
References
-
- Cameron K R, Stamminger T, Craxton M, Bodemer W, Honess R W, Fleckenstein B. The 160,000-Mr virion protein encoded at the right end of the herpesvirus saimiri genome is homologous to the 140,000-Mr membrane antigen encoded at the left end of the Epstein-Barr virus genome. J Virol. 1987;61:2063–2070. - PMC - PubMed
-
- Drouet E, Brousset P, Fares F, Icart J, Verniol C, Meggetto F, Schlaifer D, Desmorat-Coat H, Rigal-Huguet F, Niveleau A, Delsol G. High Epstein-Barr virus serum load and elevated titers of anti-ZEBRA antibodies in patients with EBV-harboring tumor cells of Hodgkin's disease. J Med Virol. 1999;57:383–389. - PubMed
-
- Gan Y J, Sullivan J L, Sixbey J W. Detection of cell-free Epstein-Barr virus DNA in serum during acute infectious mononucleosis. J Infect Dis. 1994;170:436–439. - PubMed
-
- Green M, Cacciarelli T V, Mazariegos G V, Sigurdsson L, Qu L, Rowe D T, Reyes J. Serial measurement of Epstein-Barr viral load in peripheral blood in pediatric liver transplant recipients during treatment for posttransplant lymphoproliferative disease. Transplantation. 1998;66:1641–1644. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

