Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI
- PMID: 10654941
- PMCID: PMC305580
- DOI: 10.1093/emboj/19.3.434
Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI
Abstract
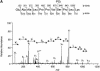
The eukaryotic translation initiation factor 4G (eIF4G) proteins play a critical role in the recruitment of the translational machinery to mRNA. The eIF4Gs are phosphoproteins. However, the location of the phosphorylation sites, how phosphorylation of these proteins is modulated and the identity of the intracellular signaling pathways regulating eIF4G phosphorylation have not been established. In this report, two-dimensional phosphopeptide mapping demonstrates that the phosphorylation state of specific eIF4GI residues is altered by serum and mitogens. Phosphopeptides resolved by this method were mapped to the C-terminal one-third of the protein. Mass spectrometry and mutational analyses identified the serum-stimulated phosphorylation sites in this region as serines 1108, 1148 and 1192. Phosphoinositide-3-kinase (PI3K) inhibitors and rapamycin, an inhibitor of the kinase FRAP/mTOR (FKBP12-rapamycin-associated protein/mammalian target of rapamycin), prevent the serum-induced phosphorylation of these residues. Finally, the phosphorylation state of N-terminally truncated eIF4GI proteins acquires resistance to kinase inhibitor treatment. These data suggest that the kinases phosphorylating serines 1108, 1148 and 1192 are not directly downstream of PI3K and FRAP/mTOR, but that the accessibility of the C-terminus to kinases is modulated by this pathway(s).
Figures





Similar articles
-
Cellular stress in xenopus kidney cells enhances the phosphorylation of eukaryotic translation initiation factor (eIF)4E and the association of eIF4F with poly(A)-binding protein.Biochem J. 1999 Sep 15;342 Pt 3(Pt 3):519-26. Biochem J. 1999. PMID: 10477262 Free PMC article.
-
Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region.Mol Cell Biol. 2000 Jan;20(2):468-77. doi: 10.1128/MCB.20.2.468-477.2000. Mol Cell Biol. 2000. PMID: 10611225 Free PMC article.
-
A direct linkage between the phosphoinositide 3-kinase-AKT signaling pathway and the mammalian target of rapamycin in mitogen-stimulated and transformed cells.Cancer Res. 2000 Jul 1;60(13):3504-13. Cancer Res. 2000. PMID: 10910062
-
Regulation of translation initiation by FRAP/mTOR.Genes Dev. 2001 Apr 1;15(7):807-26. doi: 10.1101/gad.887201. Genes Dev. 2001. PMID: 11297505 Review. No abstract available.
-
Amino acids as regulators of gene expression at the level of mRNA translation.J Nutr. 2003 Jun;133(6 Suppl 1):2046S-2051S. doi: 10.1093/jn/133.6.2046S. J Nutr. 2003. PMID: 12771363 Review.
Cited by
-
mTOR coordinates protein synthesis, mitochondrial activity and proliferation.Cell Cycle. 2015;14(4):473-80. doi: 10.4161/15384101.2014.991572. Cell Cycle. 2015. PMID: 25590164 Free PMC article. Review.
-
Chaperon-like Activation of Serum-Inducible Tryptophanyl-tRNA Synthetase Phosphorylation through Refolding as a Tool for Analysis of Clinical Samples.Transl Oncol. 2011 Dec;4(6):377-89. doi: 10.1593/tlo.11220. Epub 2011 Dec 1. Transl Oncol. 2011. PMID: 22191002 Free PMC article.
-
VPg of murine norovirus binds translation initiation factors in infected cells.Virol J. 2006 May 23;3:33. doi: 10.1186/1743-422X-3-33. Virol J. 2006. PMID: 16719923 Free PMC article.
-
mTOR signaling: implications for cancer and anticancer therapy.Br J Cancer. 2006 Jan 30;94(2):195-9. doi: 10.1038/sj.bjc.6602902. Br J Cancer. 2006. PMID: 16404421 Free PMC article. Review.
-
Translating across kingdoms: target of rapamycin promotes protein synthesis through conserved and divergent pathways in plants.J Exp Bot. 2022 Nov 15;73(20):7016-7025. doi: 10.1093/jxb/erac267. J Exp Bot. 2022. PMID: 35770874 Free PMC article. Review.
References
-
- Abraham R.T. (1998) Mammalian target of rapamycin—immunosuppressive drugs uncover a novel pathway of cytokine receptor signaling. Curr. Opin. Immunol., 10, 330–336. - PubMed
-
- Brown E.J. and Schreiber, S.L. (1996) A signaling pathway to translational control. Cell, 86, 517–520. - PubMed
-
- Browning K.S., Maia, D.M., Lax, S.R. and Ravel, J.M. (1987) Identification of a new protein synthesis initiation factor from wheat germ. J. Biol. Chem., 262, 538–541. - PubMed
-
- Chen C.A. and Okayama, H. (1988) Calcium phosphate-mediated gene transfer: a highly efficient transfection system for stably transforming cells with plasmid DNA. Biotechniques, 6, 632–638. - PubMed
-
- Craig A.W.B., Haghighat, A., Yu, A.T.K. and Sonenberg, N. (1998) Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature, 392, 520–523. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

