Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3' nontranslated region are essential for virus replication in vivo
- PMID: 10644379
- PMCID: PMC111684
- DOI: 10.1128/jvi.74.4.2046-2051.2000
Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3' nontranslated region are essential for virus replication in vivo
Abstract
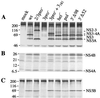
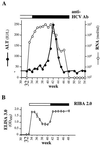
Hepatitis C virus (HCV) infection is a widespread major human health concern. Significant obstacles in the study of this virus include the absence of a reliable tissue culture system and a small-animal model. Recently, we constructed full-length HCV cDNA clones and successfully initiated HCV infection in two chimpanzees by intrahepatic injection of in vitro-transcribed RNA (A. A. Kolykhalov et al., Science 277:570-574, 1997). In order to validate potential targets for development of anti-HCV therapeutics, we constructed six mutant derivatives of this prototype infectious clone. Four clones contained point mutations ablating the activity of the NS2-3 protease, the NS3-4A serine protease, the NS3 NTPase/helicase, and the NS5B polymerase. Two additional clones contained deletions encompassing all or part of the highly conserved 98-base sequence at the 3' terminus of the HCV genome RNA. The RNA transcript from each of the six clones was injected intrahepatically into a chimpanzee. No signs of HCV infection were detected in the 8 months following the injection. Inoculation of the same animal with nonmutant RNA transcripts resulted in productive HCV infection, as evidenced by viremia, elevated serum alanine aminotransferase, and HCV-specific seroconversion. These data suggest that these four HCV-encoded enzymatic activities and the conserved 3' terminal RNA element are essential for productive replication in vivo.
Figures
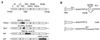
Similar articles
-
Molecular virology of hepatitis C virus: an update with respect to potential antiviral targets.Antivir Ther. 1998;3(Suppl 3):71-81. Antivir Ther. 1998. PMID: 10726057 Review.
-
Interference of HCV replication by cell penetrable human monoclonal scFv specific to NS5B polymerase.MAbs. 2014;6(5):1327-39. doi: 10.4161/mabs.29978. MAbs. 2014. PMID: 25517317 Free PMC article.
-
Hepatitis C virus NS2/3 protease regulates HCV IRES-dependent translation and NS5B RdRp activity.Arch Virol. 2009;154(9):1465-73. doi: 10.1007/s00705-009-0469-7. Epub 2009 Aug 18. Arch Virol. 2009. PMID: 19688585
-
Mutations in NS5B polymerase of hepatitis C virus: impacts on in vitro enzymatic activity and viral RNA replication in the subgenomic replicon cell culture.Virology. 2002 Jun 5;297(2):298-306. doi: 10.1006/viro.2002.1461. Virology. 2002. PMID: 12083828
-
Candidate targets for hepatitis C virus-specific antiviral therapy.Intervirology. 1997;40(5-6):378-93. doi: 10.1159/000150570. Intervirology. 1997. PMID: 9675642 Review.
Cited by
-
The Polymerase Chain Reaction: Essential for the Development of Curative Therapy for Hepatitis C.Dig Dis Sci. 2015 Aug;60(8):2232-5. doi: 10.1007/s10620-015-3748-z. Dig Dis Sci. 2015. PMID: 26072321 Free PMC article. No abstract available.
-
Effect of cell growth on hepatitis C virus (HCV) replication and a mechanism of cell confluence-based inhibition of HCV RNA and protein expression.J Virol. 2006 Feb;80(3):1181-90. doi: 10.1128/JVI.80.3.1181-1190.2006. J Virol. 2006. PMID: 16414995 Free PMC article.
-
Secondary structure of the 3' terminus of hepatitis C virus minus-strand RNA.J Virol. 2002 Aug;76(16):8058-68. doi: 10.1128/jvi.76.16.8058-8068.2002. J Virol. 2002. PMID: 12134011 Free PMC article.
-
The p7 polypeptide of hepatitis C virus is critical for infectivity and contains functionally important genotype-specific sequences.Proc Natl Acad Sci U S A. 2003 Sep 30;100(20):11646-51. doi: 10.1073/pnas.1834545100. Epub 2003 Sep 22. Proc Natl Acad Sci U S A. 2003. PMID: 14504405 Free PMC article.
-
Studying hepatitis C virus: making the best of a bad virus.J Virol. 2007 Sep;81(17):8853-67. doi: 10.1128/JVI.00753-07. Epub 2007 May 23. J Virol. 2007. PMID: 17522203 Free PMC article. Review. No abstract available.
References
-
- Beard M R, Abell G, Honda M, Carroll A, Gartland M, Clarke B, Suzuki K, Lanford R, Sangar D V, Lemon S M. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology. 1999;30:316–324. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

