Potential role for luman, the cellular homologue of herpes simplex virus VP16 (alpha gene trans-inducing factor), in herpesvirus latency
- PMID: 10623756
- PMCID: PMC111614
- DOI: 10.1128/jvi.74.2.934-943.2000
Potential role for luman, the cellular homologue of herpes simplex virus VP16 (alpha gene trans-inducing factor), in herpesvirus latency
Abstract
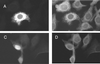
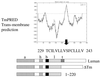
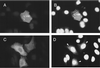
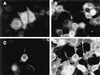
The cascade of herpes simplex virus (HSV) gene expression that results in viral replication begins with the activation of viral immediate-early (IE) genes by the virion-associated protein VP16. VP16 on its own is inefficient at associating with complexes formed on IE gene promoters and depends upon the cellular factor HCF for its activity. In this respect VP16 mimics the host basic leucine zipper (bZIP) protein Luman, which also requires HCF for activating transcription. Our objective is to explore interactions between Luman and HCF and to determine if they play a role in the biology of herpesviruses. In this report we show that in cultured cells ectopically expressed Luman was retained in the cytoplasm, where it colocalized with Calnexin, a protein normally associated with the endoplasmic reticulum (ER). Retention of Luman in the ER depends on a hydrophobic segment of the protein that probably serves as a transmembrane domain. Deletion of this domain changed the intracellular location of Luman so that most of the mutant protein was in the nucleus of cells. While HCF was present in the nucleus of most cells, in cells expressing Luman it was retained in the cytoplasm where the two proteins colocalized. This cytoplasmic association of Luman and HCF could also be demonstrated in neurons in trigeminal ganglia removed from cattle soon after death. Cells in tissue culture that expressed Luman, but not a mutant form of the protein that fails to bind HCF, were resistant to a productive infection with HSV type 1 (HSV-1). We hypothesize that similar Luman-HCF interactions in sensory neurons in trigeminal ganglia result in the suppression of viral replication and the establishment of latency. Interestingly, Luman could activate the promoters of IE110 and LAT, two genes that are critical for reactivation of HSV-1 from latency. This suggests a role for Luman in the reactivation process as well.
Figures



Similar articles
-
Zhangfei: a second cellular protein interacts with herpes simplex virus accessory factor HCF in a manner similar to Luman and VP16.Nucleic Acids Res. 2000 Jun 15;28(12):2446-54. doi: 10.1093/nar/28.12.2446. Nucleic Acids Res. 2000. PMID: 10871379 Free PMC article.
-
The herpesvirus transactivator VP16 mimics a human basic domain leucine zipper protein, luman, in its interaction with HCF.J Virol. 1998 Aug;72(8):6291-7. doi: 10.1128/JVI.72.8.6291-6297.1998. J Virol. 1998. PMID: 9658067 Free PMC article.
-
Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor.Mol Cell Biol. 1997 Sep;17(9):5117-26. doi: 10.1128/MCB.17.9.5117. Mol Cell Biol. 1997. PMID: 9271389 Free PMC article.
-
Host factors associated with either VP16 or VP16-induced complex differentially affect HSV-1 lytic infection.Rev Med Virol. 2022 Nov;32(6):e2394. doi: 10.1002/rmv.2394. Epub 2022 Sep 7. Rev Med Virol. 2022. PMID: 36069169 Free PMC article. Review.
-
Early expression of herpes simplex virus (HSV) proteins and reactivation of latent infection.Folia Microbiol (Praha). 2000;45(1):7-28. doi: 10.1007/BF02817445. Folia Microbiol (Praha). 2000. PMID: 11200675 Review.
Cited by
-
Recruitment of the transcriptional coactivator HCF-1 to viral immediate-early promoters during initiation of reactivation from latency of herpes simplex virus type 1.J Virol. 2009 Sep;83(18):9591-5. doi: 10.1128/JVI.01115-09. Epub 2009 Jul 1. J Virol. 2009. PMID: 19570863 Free PMC article.
-
Tegument protein control of latent herpesvirus establishment and animation.Herpesviridae. 2011 Feb 8;2(1):3. doi: 10.1186/2042-4280-2-3. Herpesviridae. 2011. PMID: 21429246 Free PMC article.
-
Gene array analysis reveals changes in peripheral nervous system gene expression following stimuli that result in reactivation of latent herpes simplex virus type 1: induction of transcription factor Bcl-3.J Virol. 2001 Oct;75(20):9909-17. doi: 10.1128/JVI.75.20.9909-9917.2001. J Virol. 2001. PMID: 11559823 Free PMC article.
-
Stabilization but not the transcriptional activity of herpes simplex virus VP16-induced complexes is evolutionarily conserved among HCF family members.J Virol. 2001 Dec;75(24):12402-11. doi: 10.1128/JVI.75.24.12402-12411.2001. J Virol. 2001. PMID: 11711630 Free PMC article.
-
Insights from the crystal structure of the chicken CREB3 bZIP suggest that members of the CREB3 subfamily transcription factors may be activated in response to oxidative stress.Protein Sci. 2019 Apr;28(4):779-787. doi: 10.1002/pro.3573. Epub 2019 Feb 6. Protein Sci. 2019. PMID: 30653278 Free PMC article.
References
-
- Bloom D C, Stevens J G, Hill J M, Tran R K. Mutagenesis of a cAMP response element within the latency-associated transcript promoter of HSV-1 reduces adrenergic reactivation. Virology. 1997;236:202–207. - PubMed
-
- Brown M S, Goldstein J L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

