In vitro evidence for the interaction of tRNA(3)(Lys) with U3 during the first strand transfer of HIV-1 reverse transcription
- PMID: 10606665
- PMCID: PMC102502
- DOI: 10.1093/nar/28.2.634
In vitro evidence for the interaction of tRNA(3)(Lys) with U3 during the first strand transfer of HIV-1 reverse transcription
Abstract
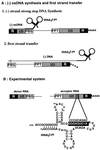
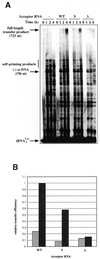
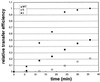
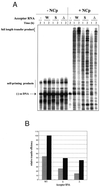
Over the course of its evolution, HIV-1 has taken maximum advantage of its tRNA(3)(Lys)primer by utilizing it in several steps of reverse transcription. Here, we have identified a conserved nonanucleotide sequence in the U3 region of HIV-1 RNA that is complementary to the anticodon stem of tRNA(3)(Lys). In order to test its possible role in the first strand transfer reaction, we applied an assay using a donor RNA corresponding to the 5'-part and an acceptor RNA spanning the 3'-part of HIV-1 RNA. In addition, we constructed two acceptor RNAs in which the nonanucleotide sequence complementary to tRNA(3)(Lys)was either substituted (S) or deleted (Delta). We used either natural tRNA(3)(Lys)or an 18 nt DNA as primer and measured the efficiency of (-) strand strong stop DNA transfer in the presence of wild-type, S or Delta acceptor RNA. Mutations in U3 did not decrease the transfer efficiency when reverse transcription was primed with the 18mer DNA. However, they significantly reduced the strand transfer efficiency in the tRNA(3)(Lys)-primed reactions. This reduction was also observed in the presence of nucleocapsid protein. These results suggest that tRNA(3)(Lys)increases (-) strand strong stop transfer by interacting with the U3 region of the genomic RNA. Sequence comparisons suggest that such long range interactions also exist in other lentiviruses.
Figures


Similar articles
-
Role of post-transcriptional modifications of primer tRNALys,3 in the fidelity and efficacy of plus strand DNA transfer during HIV-1 reverse transcription.J Biol Chem. 1999 Feb 12;274(7):4412-20. doi: 10.1074/jbc.274.7.4412. J Biol Chem. 1999. PMID: 9933645
-
Comparison of deoxyoligonucleotide and tRNA(Lys-3) as primers in an endogenous human immunodeficiency virus-1 in vitro reverse transcription/template-switching reaction.J Biol Chem. 1994 May 20;269(20):14672-80. J Biol Chem. 1994. PMID: 7514178
-
The HIV plus-strand transfer reaction: determination of replication-competent intermediates and identification of a novel lentiviral element, the primer over-extension sequence.J Mol Biol. 2002 Jan 18;315(3):311-23. doi: 10.1006/jmbi.2001.5205. J Mol Biol. 2002. PMID: 11786014
-
The selective packaging and annealing of primer tRNALys3 in HIV-1.Curr HIV Res. 2004 Apr;2(2):163-75. doi: 10.2174/1570162043484988. Curr HIV Res. 2004. PMID: 15078180 Review.
-
Initiation of HIV-1 reverse transcription and functional role of nucleocapsid-mediated tRNA/viral genome interactions.Virus Res. 2012 Nov;169(2):324-39. doi: 10.1016/j.virusres.2012.06.006. Epub 2012 Jun 18. Virus Res. 2012. PMID: 22721779 Review.
Cited by
-
Molecular mimicry of human tRNALys anti-codon domain by HIV-1 RNA genome facilitates tRNA primer annealing.RNA. 2013 Feb;19(2):219-29. doi: 10.1261/rna.036681.112. Epub 2012 Dec 21. RNA. 2013. PMID: 23264568 Free PMC article.
-
A sequence similar to tRNA 3 Lys gene is embedded in HIV-1 U3-R and promotes minus-strand transfer.Nat Struct Mol Biol. 2010 Jan;17(1):83-9. doi: 10.1038/nsmb.1687. Epub 2009 Dec 6. Nat Struct Mol Biol. 2010. PMID: 19966801 Free PMC article.
-
Vif is a RNA chaperone that could temporally regulate RNA dimerization and the early steps of HIV-1 reverse transcription.Nucleic Acids Res. 2007;35(15):5141-53. doi: 10.1093/nar/gkm542. Epub 2007 Jul 26. Nucleic Acids Res. 2007. PMID: 17660191 Free PMC article.
-
Subtle alterations of the native zinc finger structures have dramatic effects on the nucleic acid chaperone activity of human immunodeficiency virus type 1 nucleocapsid protein.J Virol. 2002 May;76(9):4370-8. doi: 10.1128/jvi.76.9.4370-4378.2002. J Virol. 2002. PMID: 11932404 Free PMC article.
-
Nucleotides within the anticodon stem are important for optimal use of tRNA(Lys,3) as the primer for HIV-1 reverse transcription.Virology. 2007 Jul 20;364(1):169-77. doi: 10.1016/j.virol.2007.02.010. Epub 2007 Mar 21. Virology. 2007. PMID: 17368706 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

