Invasion by Toxoplasma gondii establishes a moving junction that selectively excludes host cell plasma membrane proteins on the basis of their membrane anchoring
- PMID: 10601353
- PMCID: PMC2195726
- DOI: 10.1084/jem.190.12.1783
Invasion by Toxoplasma gondii establishes a moving junction that selectively excludes host cell plasma membrane proteins on the basis of their membrane anchoring
Abstract
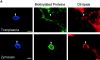
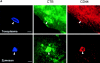
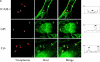
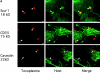
The protozoan parasite Toxoplasma gondii actively penetrates its host cell by squeezing through a moving junction that forms between the host cell plasma membrane and the parasite. During invasion, this junction selectively controls internalization of host cell plasma membrane components into the parasite-containing vacuole. Membrane lipids flowed past the junction, as shown by the presence of the glycosphingolipid G(M1) and the cationic lipid label 1. 1'-dihexadecyl-3-3'-3-3'-tetramethylindocarbocyanine (DiIC(16)). Glycosylphosphatidylinositol (GPI)-anchored surface proteins, such as Sca-1 and CD55, were also readily incorporated into the parasitophorous vacuole (PV). In contrast, host cell transmembrane proteins, including CD44, Na(+)/K(+) ATPase, and beta1-integrin, were excluded from the vacuole. To eliminate potential differences in sorting due to the extracellular domains, parasite invasion was examined in host cells transfected with recombinant forms of intercellular adhesion molecule 1 (ICAM-1, CD54) that differed in their mechanism of membrane anchoring. Wild-type ICAM-1, which contains a transmembrane domain, was excluded from the PV, whereas both GPI-anchored ICAM-1 and a mutant of ICAM-1 missing the cytoplasmic tail (ICAM-1-Cyt(-)) were readily incorporated into the PV membrane. Our results demonstrate that during host cell invasion, Toxoplasma selectively excludes host cell transmembrane proteins at the moving junction by a mechanism that depends on their anchoring in the membrane, thereby creating a nonfusigenic compartment.
Figures








Similar articles
-
Toxoplasma gondii resides in a vacuole that avoids fusion with host cell endocytic and exocytic vesicular trafficking pathways.Exp Parasitol. 1999 Jun;92(2):87-99. doi: 10.1006/expr.1999.4412. Exp Parasitol. 1999. PMID: 10366534
-
Interactions between Toxoplasma gondii and its mammalian host cells.Semin Cell Biol. 1993 Oct;4(5):335-44. doi: 10.1006/scel.1993.1040. Semin Cell Biol. 1993. PMID: 8257785 Review.
-
Microneme proteins: structural and functional requirements to promote adhesion and invasion by the apicomplexan parasite Toxoplasma gondii.Int J Parasitol. 2001 Oct;31(12):1293-302. doi: 10.1016/s0020-7519(01)00257-0. Int J Parasitol. 2001. PMID: 11566297 Review.
-
The role of the cytoskeleton in host cell invasion by Toxoplasma gondii.Behring Inst Mitt. 1997 Mar;(99):90-6. Behring Inst Mitt. 1997. PMID: 9303207 Review.
-
Host but not parasite cholesterol controls Toxoplasma cell entry by modulating organelle discharge.Mol Biol Cell. 2003 Sep;14(9):3804-20. doi: 10.1091/mbc.e02-12-0830. Epub 2003 May 29. Mol Biol Cell. 2003. PMID: 12972565 Free PMC article.
Cited by
-
Host-cell lipid rafts: a safe door for micro-organisms?Biol Cell. 2010 Apr 6;102(7):391-407. doi: 10.1042/BC20090138. Biol Cell. 2010. PMID: 20377525 Free PMC article. Review.
-
An apical protein, Pcr2, is required for persistent movement by the human parasite Toxoplasma gondii.PLoS Pathog. 2022 Aug 22;18(8):e1010776. doi: 10.1371/journal.ppat.1010776. eCollection 2022 Aug. PLoS Pathog. 2022. PMID: 35994509 Free PMC article.
-
Interplay Between Toxoplasma gondii, Autophagy, and Autophagy Proteins.Front Cell Infect Microbiol. 2019 May 1;9:139. doi: 10.3389/fcimb.2019.00139. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31119109 Free PMC article. Review.
-
Toxoplasma rhoptries: unique secretory organelles and source of promising vaccine proteins for immunoprevention of toxoplasmosis.J Biomed Biotechnol. 2008;2008:632424. doi: 10.1155/2008/632424. J Biomed Biotechnol. 2008. PMID: 18670609 Free PMC article. Review.
-
Unraveling the Cave: A Seventy-Year Journey into the Caveolar Network, Cellular Signaling, and Human Disease.Cells. 2023 Nov 22;12(23):2680. doi: 10.3390/cells12232680. Cells. 2023. PMID: 38067108 Free PMC article. Review.
References
-
- Aikawa M., Sterling C.R. Intracellular Parasitic Protozoa 1974. Academic Press, Inc; New York: pp. 76
-
- Dubey J.P., Beattie C.P. Toxoplasmosis of Animals and Man 1988. CRC Press; Boca Raton, FL: pp. 220
-
- Morisaki J.H., Heuser J.E., Sibley L.D. Invasion of Toxoplasma gondii occurs by active penetration of the host cell. J. Cell Sci. 1995;108:2457–2464. - PubMed
-
- Dobrowolski J.M., Sibley L.D. Toxoplasma invasion of mammalian cells is powered by the actin cytoskeleton of the parasite. Cell. 1996;84:933–939. - PubMed
-
- Mordue D.G., Håkansson S., Niesman I., Sibley L.D. Toxoplasma gondii resides in a vacuole that avoids fusion with host cell endocytic and exocytic vesicular trafficking pathways. Exp. Parasitol. 1999;92:87–99. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

