Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase
- PMID: 10601033
- PMCID: PMC317176
- DOI: 10.1101/gad.13.23.3070
Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase
Abstract
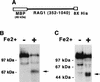
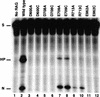
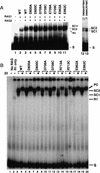
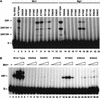
The RAG1 and RAG2 proteins collaborate to initiate V(D)J recombination by binding recombination signal sequences (RSSs) and making a double-strand break between the RSS and adjacent coding DNA. Like the reactions of their biochemical cousins, the bacterial transposases and retroviral integrases, cleavage by the RAG proteins requires a divalent metal ion but does not involve a covalent protein/DNA intermediate. In the transposase/integrase family, a triplet of acidic residues, commonly called a DDE motif, is often found to coordinate the metal ion used for catalysis. We show here that mutations in each of three acidic residues in RAG1 result in mutant derivatives that can bind the RSS but whose ability to catalyze either of the two chemical steps of V(D)J cleavage (nicking and hairpin formation) is severely impaired. Because both chemical steps are affected by the same mutations, a single active site appears responsible for both reactions. Two independent lines of evidence demonstrate that at least two of these acidic residues are directly involved in coordinating a divalent metal ion: The substitution of Cys for Asp allows rescue of some catalytic function, whereas an alanine substitution is no longer subject to iron-induced hydroxyl radical cleavage. Our results support a model in which the RAG1 protein contains the active site of the V(D)J recombinase and are interpreted in light of predictions about the structure of RAG1.
Figures


Similar articles
-
Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination.Genes Dev. 1999 Dec 1;13(23):3059-69. doi: 10.1101/gad.13.23.3059. Genes Dev. 1999. PMID: 10601032 Free PMC article.
-
Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex.Mol Cell. 2000 Jan;5(1):97-107. doi: 10.1016/s1097-2765(00)80406-2. Mol Cell. 2000. PMID: 10678172
-
The DDE motif in RAG-1 is contributed in trans to a single active site that catalyzes the nicking and transesterification steps of V(D)J recombination.Mol Cell Biol. 2001 Jan;21(2):449-58. doi: 10.1128/MCB.21.2.449-458.2001. Mol Cell Biol. 2001. PMID: 11134333 Free PMC article.
-
The RAG proteins and V(D)J recombination: complexes, ends, and transposition.Annu Rev Immunol. 2000;18:495-527. doi: 10.1146/annurev.immunol.18.1.495. Annu Rev Immunol. 2000. PMID: 10837067 Review.
-
RAG1 and RAG2 in V(D)J recombination and transposition.Immunol Res. 2001;23(1):23-39. doi: 10.1385/IR:23:1:23. Immunol Res. 2001. PMID: 11417858 Review.
Cited by
-
Higher-order looping and nuclear organization of Tcra facilitate targeted rag cleavage and regulated rearrangement in recombination centers.Cell Rep. 2013 Feb 21;3(2):359-70. doi: 10.1016/j.celrep.2013.01.024. Epub 2013 Feb 14. Cell Rep. 2013. PMID: 23416051 Free PMC article.
-
Novel molecular mechanism for generating NK-cell fitness and memory.Eur J Immunol. 2015 Jul;45(7):1906-15. doi: 10.1002/eji.201445339. Epub 2015 Jun 24. Eur J Immunol. 2015. PMID: 26018782 Free PMC article. Review.
-
Identification of a divalent metal cation binding site in herpes simplex virus 1 (HSV-1) ICP8 required for HSV replication.J Virol. 2012 Jun;86(12):6825-34. doi: 10.1128/JVI.00374-12. Epub 2012 Apr 4. J Virol. 2012. PMID: 22491472 Free PMC article.
-
The origins of the Rag genes--from transposition to V(D)J recombination.Semin Immunol. 2010 Feb;22(1):10-6. doi: 10.1016/j.smim.2009.11.004. Epub 2009 Dec 9. Semin Immunol. 2010. PMID: 20004590 Free PMC article. Review.
-
Base flipping in V(D)J recombination: insights into the mechanism of hairpin formation, the 12/23 rule, and the coordination of double-strand breaks.Mol Cell Biol. 2009 Nov;29(21):5889-99. doi: 10.1128/MCB.00187-09. Epub 2009 Aug 31. Mol Cell Biol. 2009. PMID: 19720743 Free PMC article.
References
-
- Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. - PubMed
-
- Allingham JS, Pribil PA, Haniford DB. All three residues of the Tn 10 transposase DDE catalytic triad function in divalent metal ion binding. J Mol Biol. 1999;289:1195–1206. - PubMed
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: Greene Publishing Associates and Wiley-Interscience; 1989.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
