Methylation-dependent gene silencing induced by interleukin 1beta via nitric oxide production
- PMID: 10587350
- PMCID: PMC2195731
- DOI: 10.1084/jem.190.11.1595
Methylation-dependent gene silencing induced by interleukin 1beta via nitric oxide production
Abstract
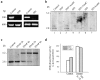
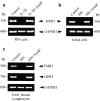
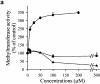
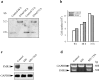
Interleukin (IL)-1beta is a pleiotropic cytokine implicated in a variety of activities, including damage of insulin-producing cells, brain injury, or neuromodulatory responses. Many of these effects are mediated by nitric oxide (NO) produced by the induction of NO synthase (iNOS) expression. We report here that IL-1beta provokes a marked repression of genes, such as fragile X mental retardation 1 (FMR1) and hypoxanthine phosphoribosyltransferase (HPRT), having a CpG island in their promoter region. This effect can be fully prevented by iNOS inhibitors and is dependent on DNA methylation. NO donors also cause FMR1 and HPRT gene silencing. NO-induced methylation of FMR1 CpG island can be reverted by demethylating agents which, in turn, produce the recovery of gene expression. The effects of IL-1beta and NO appear to be exerted through activation of DNA methyltransferase (DNA MeTase). Although exposure of the cells to NO does not increase DNA MeTase gene expression, the activity of the enzyme selectively increases when NO is applied directly on a nuclear protein extract. These findings reveal a previously unknown effect of IL-1beta and NO on gene expression, and demonstrate a novel pathway for gene silencing based on activation of DNA MeTase by NO and acute modification of CpG island methylation.
Figures

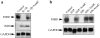
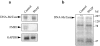


Similar articles
-
Alveolar macrophages autoregulate IL-1 and IL-6 production by endogenous nitric oxide.Am J Respir Cell Mol Biol. 1996 Mar;14(3):272-8. doi: 10.1165/ajrcmb.14.3.8845178. Am J Respir Cell Mol Biol. 1996. PMID: 8845178
-
[Methylation and expression of the FMR1 gene].Rev Neurol. 2001 Oct;33 Suppl 1:S57-62. Rev Neurol. 2001. PMID: 12447821 Spanish.
-
Suppressive effects of a selective inducible nitric oxide synthase (iNOS) inhibitor on pancreatic beta-cell dysfunction.Diabetologia. 2003 Sep;46(9):1228-33. doi: 10.1007/s00125-003-1173-x. Epub 2003 Jul 24. Diabetologia. 2003. PMID: 12898012
-
FMR1 silencing and the signals to chromatin: a unified model of transcriptional regulation.Biochem Biophys Res Commun. 2002 Jul 19;295(3):575-81. doi: 10.1016/s0006-291x(02)00682-4. Biochem Biophys Res Commun. 2002. PMID: 12099676 Review.
-
CpG islands.EXS. 1993;64:169-85. doi: 10.1007/978-3-0348-9118-9_8. EXS. 1993. PMID: 8418949 Review. No abstract available.
Cited by
-
How does the social environment 'get into the mind'? Epigenetics at the intersection of social and psychiatric epidemiology.Soc Sci Med. 2012 Jan;74(1):67-74. doi: 10.1016/j.socscimed.2011.09.036. Epub 2011 Nov 6. Soc Sci Med. 2012. PMID: 22119520 Free PMC article. Review.
-
Epigenetic analysis identifies factors driving racial disparity in prostate cancer.Cancer Rep (Hoboken). 2019 Apr;2(2):e1153. doi: 10.1002/cnr2.1153. Epub 2018 Dec 13. Cancer Rep (Hoboken). 2019. PMID: 32721098 Free PMC article.
-
Application progress of liquid biopsy in gastric cancer.Front Oncol. 2022 Sep 15;12:969866. doi: 10.3389/fonc.2022.969866. eCollection 2022. Front Oncol. 2022. PMID: 36185234 Free PMC article. Review.
-
Helicobacter pylori infection promotes methylation and silencing of trefoil factor 2, leading to gastric tumor development in mice and humans.Gastroenterology. 2010 Dec;139(6):2005-17. doi: 10.1053/j.gastro.2010.08.043. Epub 2010 Aug 27. Gastroenterology. 2010. PMID: 20801119 Free PMC article.
-
Epigenetic alterations in human prostate cancers.Endocrinology. 2009 Sep;150(9):3991-4002. doi: 10.1210/en.2009-0573. Epub 2009 Jun 11. Endocrinology. 2009. PMID: 19520778 Free PMC article. Review.
References
-
- Bird A.P. CpG islands and the function of DNA methylation. Nature. 1986;321:209–213. - PubMed
-
- Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. - PubMed
-
- Tate P.H., Bird A.P. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr. Opin. Genet. Dev. 1993;3:226–231. - PubMed
-
- Eden S., Cedar H. Role of DNA methylation in the regulation of transcription. Curr. Opin. Genet. Dev. 1994;4:255–259. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

